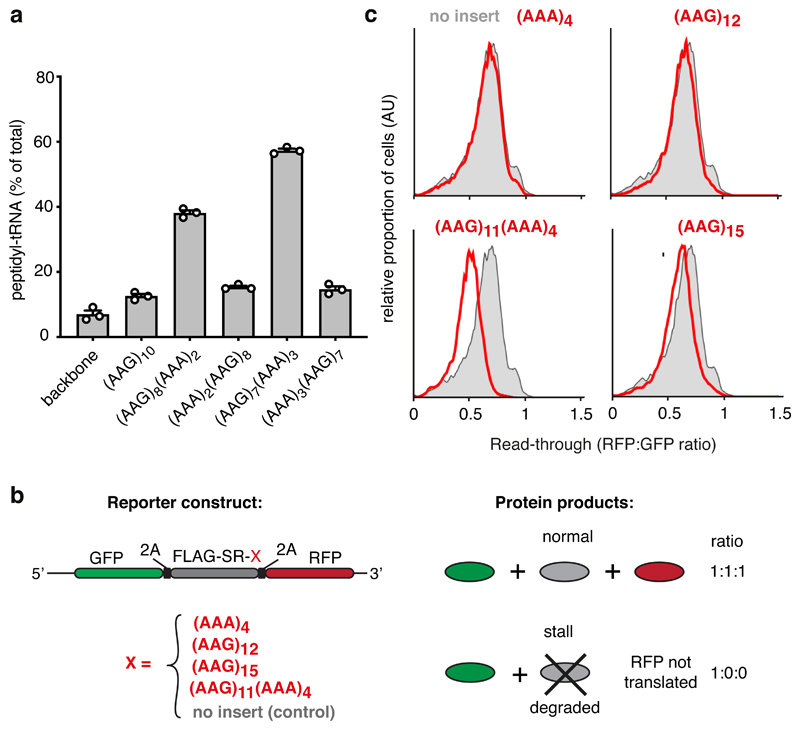
Fig. 6. Coincidence detection of the nascent chain and mRNA mediate stalling.
(A) Iterated combinations of AAG and AAA codons inserted near the end of an open reading frame were analyzed for stalling by in vitro translation. The proportion of stalled product was assessed by the relative amount of peptidyl-tRNA versus full length terminated polypeptide and plotted as % peptidyl-tRNA. A construct lacking the AAG/AAA insert is shown for comparison (‘backbone’). The mean ± SEM (n=3) together with individual data points from independent experiments are plotted. (B) Schematic diagram of the dual-color reporter construct for detection of terminal stalling by flow cytometry. “2A” indicates the viral P2A sequence which causes skipping of peptide bond formation without interrupting elongation. If a test sequence inserted downstream of the FLAG-SR region causes terminal stalling, production of GFP is unaffected while RFP is not produced. Thus, the RFP:GFP ratio will be less than 1 and serves as a quantitative measure of terminal stalling. (C) Iterated combinations of AAG and AAA codons were analyzed for stalling in HEK293T cells using a dual color reporter in which the test sequence of interest (red text) is inserted between an N-terminal GFP and C-terminal RFP. Shown are histograms from flow cytometry analysis of the RFP:GFP ratio as a measure of ribosome read-through of the test sequence (red traces) relative to that seen in the absence of an insert (grey trace). Gating strategy for the histogram is shown in Source Data online.