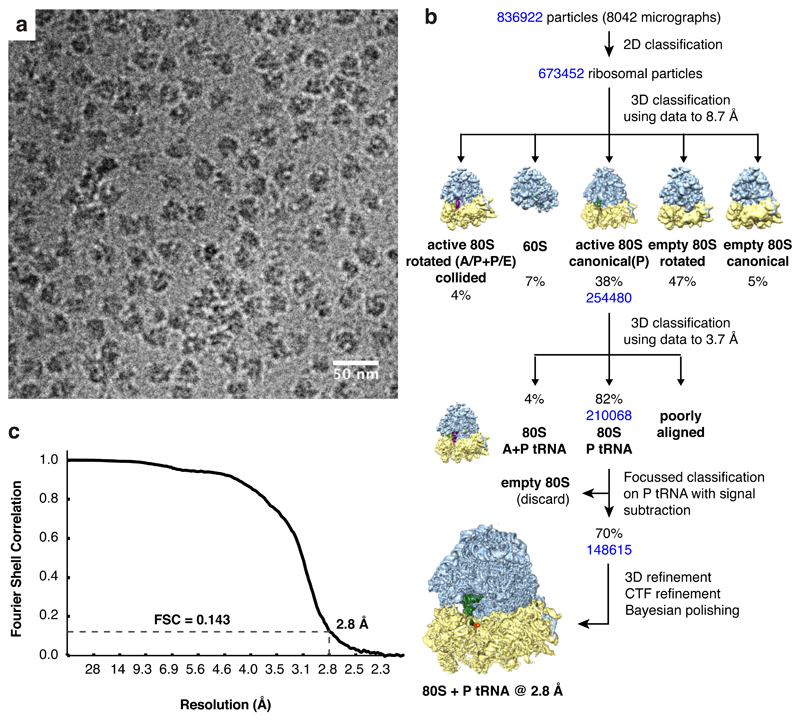
Extended Data Fig. 2. Cryo-EM analysis of ribosomes stalled on poly(A).
(A) Representative micrograph of poly(A)-stalled ribosomes used for single particle analysis. (B) Data processing scheme used for structure determination in Relion 3.0. 3D classification reveals that ~90% of active ribosomes are in the canonical state with P/P tRNA while ~10% are seen in the rotated state with A/P and P/E hybrid state tRNAs. The majority of the rotated state ribosomes also contain density for a preceding ribosome and therefore represent ribosomes that have collided with a poly(A)-stalled ribosome. (C) Fourier shell correlation (FSC) curve of the final map illustrating an overall resolution of 2.8 Å.