Abstract
Background
Primary spontaneous pneumothorax (PSP) is a disease characterized by the accumulation of air in the pleural space between the lung and thoracic wall. It is more common in young, tall, thin, and asthenic men. A family history was reported for approximately 11.5% of individuals admitted with PSP. The literature has reported cases diagnosed with familial PSP, who have no manifestations of Birt–Hogg–Dubé (BHD) syndrome but mutations in different exons of the Folliculin (FLCN) gene. The aim of this study is to present a Turkish family in which 13 members from three generations of the same family developed recurrent isolated spontaneous pneumothorax with a novel mutation in the FLCN.
Methods
A male proband was diagnosed with spontaneous pneumothorax in the emergency department of the University of Health Sciences Haydarpasa Numune Training and Research Center, Istanbul, Turkey. His 12 relatives from three generations diagnosed with PSP, as revealed by his family history, were invited to the hospital to give blood samples for mutation analysis. The Sanger sequence data of FLCN were analyzed on the ENSEMBL website using SeqScape 3 and Codon Aligner software.
Results
A novel heterozygous mutation c. 1273C>T (p.Gln425Ter) was detected in exon 11 of the FLCN, which caused PSP in the proband and his 12 relatives tested using Sanger sequencing.
Conclusion
We found that a heterozygous mutation in exon 11 of FLCN c. 1273C>T (p.Gln425Ter), which was identified for the first time in our study, might cause isolated familial spontaneous pneumothorax.
Keywords: FLCN, mutation, pneumothorax
We found that a heterozygous mutation in exon 11 of FLCN c. 1273C>T (p.Gln425Ter), which was identified for the first time in our study, might cause isolated familial spontaneous pneumothorax.
1. INTRODUCTION
Primary spontaneous pneumothorax (PSP) is a disease in which air is accumulated in the pleural space of individuals without underlying lung disease, not due to trauma and surgical intervention (Chiu & Garcia, 2006). It is more common in tall, thin, and asthenic men aged between 10 and 30. The incidence of PSP was 7.4–18/100,000 in men and 1.2–6/100,000 in women (Chiu & Garcia, 2006; Sahn & Heffner, 2000). PSP is considered to be caused by a rupture of subpleural blebs or bullae (Graham, Nolasco, Peterlin, & Garcia, 2005); however, the formation mechanism of blebs and bullae has not been clearly explained. A family history was reported for approximately 11.5% of individuals admitted with PSP (Graham et al., 2005). Coexisting monogenic disorders in PSP include alpha‐1 antitrypsin deficiency, Marfan syndrome, Ehlers‐Danlos syndrome, homocystinuria, and Birt–Hogg–Dubé (BHD) syndrome (Xing et al., 2017). The literature has reported cases with multiple PSP patients in the same pedigree who do not have the manifestations of these genetic disorders (Abolnik, Lossos, Zlotogora, & Brauer, 1991; Delaney, Gale, & Walker, 1974; Engdahl & Gershan, 1998; Gibson, 1977; Lenler‐Petersen, Grunnet, Jespersen, & Jaeger, 1990; Morrison, Lowry, & Nevin, 1998; Wilson & Aylsworth, 1979; Yamada, Takeda, Hayashi, & Shimizu, 2003).
Familial PSP was first defined by Faber in 1921 (Xing et al., 2017). Research has shown that the genetic inheritance of isolated PSP may be autosomal dominant, autosomal recessive, or X‐linked recessive (Fröhlich et al., 2008). The folliculin (FLCN) gene, a tumor suppressor gene localized in the short arm of the chromosome (17p11.2), was first identified in 2001 and reported to be associated with PSP, fibrofolliculoma, pulmonary cyst, and Birt–Hogg–Dubé Syndrome‐associated kidney cysts (Graham et al., 2005). Mutations in different exons of FLCN have recently been reported in patients who had no clinical manifestations of α1‐antitrypsin deficiency, connective tissue disorders, or Birt–Hogg–Dubé Syndrome but were only diagnosed with PSP (Fröhlich et al., 2008).
In the present study, we identified a novel and previously not reported mutation in FLCN in 13 individuals who had no clinical manifestations of α1‐antitrypsin deficiency, connective tissue disorders (e.g., Marfan syndrome or Ehlers‐Danlos syndrome), or BHD syndrome.
2. METHODS
2.1. Blood sampling
A 60‐year‐old male proband of Turkish origin (II‐5) was diagnosed with spontaneous pneumothorax (Figure 1) in the emergency department of the University of Health Sciences Haydarpasa Numune Training and Research Center, Istanbul. His 12 relatives from three generations diagnosed with PSP, as revealed by his family history, were invited to the hospital. We evaluated when they had a pneumothorax attack, whether it was due to trauma or iatrogenic cause, and which therapies were used as well as the results of thoracic computed tomography. Blood samples were collected from affected individuals for FLCN mutation analysis.
Figure 1.
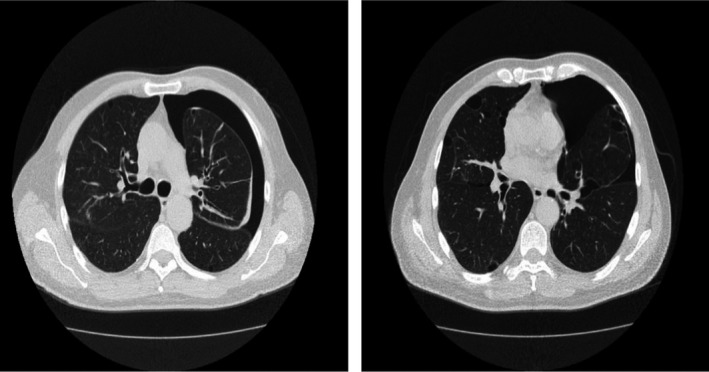
Chest high‐resolution computed tomography of the proband, who experienced recurrent episodes of PSP. Left‐sided PSP and bilateral and multiple bullae are seen as clear. PSP, primary spontaneous pneumothorax
2.2. Mutation analysis
Peripheral blood samples were collected into EDTA anticoagulant tubes and stored at +4°C until DNA isolation was performed. Genomic DNA was extracted from the blood samples of the proband case and family members in accordance with the instructions of the manufacturer. DNA isolation of the samples was performed using an automatic instrument kit (Qiagen, EZ1 Advanced XL). NanoDrop ND‐2000c (Thermo Fisher, Inc.) was used to quantify and assess the purity and concentration of isolated DNA samples. The DNA samples were stored at −20°C until the polymerase chain reaction (PCR) stage. The final amount of each PCR mixture was set to be 25 µl for the amplification of the FLCN 4‐14 exons. DreamTaq PCR Master Mix (2X) (Thermo Scientific) was used in the PCR and the primer pair shown in Table 1 was used for FLCN (van Steensel et al., 2007). 100 μmol primers were diluted to 10 μmol and 0.5 µl was used per reaction. The following touchdown PCR conditions were used: 94°C for 5 minutes followed by 16 cycles of 94°C for 30 s, 68°C for 30 s, 72°C for 1 minute; then 25 cycles of 94°C for 30 s, 55°C for 30 s, 72°C for 1 minute, and 72°C for 10 minutes. The PCR products were electrophoresed on a 2% agarose gel followed by PCR purification by ExoSAP (GML). The DNA sequencing reaction was performed using the BigDye Version 1.1 (Life Technologies) and the sequencing was performed using the 3500 Genetic Analyzer (Applied Biosystems). The sequence data were analyzed on the ENSEMBL website using SeqScape™ Software v3.0 and Codon Aligner Software 3.0 with the ENST00000285071 transcript. We also used Jmol Software 13.0 program to show whether any conformational change in protein structure of the FLCN ("Jmol: an open‐source Java viewer for chemical structures in D.").
Table 1.
List of primer sequences for polymerase chain reaction amplification
| Primer name | Primer sequences 5’→3’ |
|---|---|
| FLCN4F | 5'‐AGG TGC TCC CTG TGC TCC AG‐3' |
| FLCN4R | 5'‐CCG TCC ACT GCT CTC AGG TC‐3' |
| FLCN5F | 5'‐CCG AGC TCA GAT TTG CAT AAA CC‐3' |
| FLCN5R | 5'‐CCT GCC TCC CTG TGC AAT G‐3' |
| FLCN6F | 5'‐TGA TTT GTG CCA GCT GAC TCT G‐3' |
| FLCN6R | 5'‐CCA GGC CTC AAC CTC AGC AC‐3' |
| FLCN7F | 5'‐CCT GGA GTT GGC TGT GAA CG‐3' |
| FLCN7R | 5'‐TCC CAA ATC CAT GGA CAA GC‐3' |
| FLCN8F | 5'‐GTT GTG CCC TGC TGG TGT TC‐3' |
| FLCN8R | 5'‐TTC CCT CCC TCA GCG ATT CC‐3' |
| FLCN9F | 5'‐GGC CGC AGC CAG GAA TCT/AC3' |
| FLCN9R | 5'‐GTG GAG GGT CCA GAG GCA AG‐3' |
| FLCN10F | 5'‐CAC CCG CCT CCC TGA GAA G‐3' |
| FLCN10R | 5'‐CCA GTG GAG ACC GTG TGG TG‐3' |
| FLCN11F | 5'‐GGT TCC ACT TTG GGC CTG AG‐3' |
| FLCN11R | 5'‐AGG AGG CGT GTG GGG TTT G‐3' |
| FLCN12F | 5'‐CTA GCG CAG GGG AGG TGA GG‐3' |
| FLCN13R | 5'‐ACG GCC CAG CTC CTC TTT TG‐3' |
| FLCN14F | 5'‐CCG TGT CAC CCC TGG TTG G‐3' |
| FLCN14R | 5'‐TGC TGG GAC ACA GCT CCT TC‐3' |
3. RESULTS
In our study, PSP was detected in 13 cases, including the proband. We found that 11 of the family members underwent invasive treatment and two family members underwent conservative treatment due to PSP. Figure 2 shows the pedigree of the family members included in the study. The skin examination showed that none of the patients had fibrofolliculoma skin lesions. The ultrasound imaging of the urinary system showed that they had no renal mass. Additionally, none of the patients had clinical manifestations of BHD syndrome or a connective tissue disease such as Marfan syndrome and Ehlers‐Danlos syndrome. In the patients, α1‐antitrypsin deficiency was excluded. Among 13 patients included in the study, 77% (n = 10) were female and 23% (n = 3) were male. The patients had a total of 32 pneumothorax attacks. The mean age at the first pneumothorax attack was 40.23 years and the mean BMI was 25.55 kg/m2. The median number of episodes was 2.46. All 13 patients had bilateral and multiple air cysts (bullae and blebs) on thorax CT images. All patients had normal respiratory function tests. None of the patients had interstitial lung diseases or diffuse parenchymal lung diseases such as bronchiectasis and tuberculosis.
Figure 2.
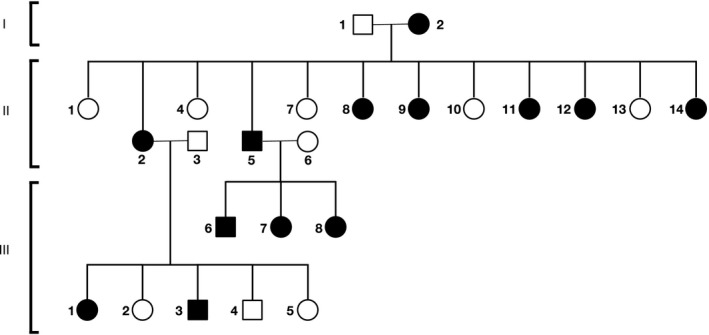
Pedigree of the Turkish family with primary spontaneous pneumothorax. Circles are females, squares are males, affected individuals are highlighted in black. Roman numbers indicate generations and Arabic numbers indicate individuals. The index patient is II‐5
The proband's mother (I‐2) underwent chest tube drainage when she was 68 years old due to left PSP. She had no recurrence so far. In the second generation, six of 11 sisters (II‐2, 8, 9, 11, 12, and 14) had a history of PSP. In the third generation, three children (III‐6, 7, and 8) of the proband and two children (III‐1 and 3) of one of the six sisters (II‐2) had a history of PSP. There was no consanguineous marriage in the first generation and the parents of the third generation had no ties of kinship. Table 2 displays the clinical data of 13 family members with a history of PSP. None of the family members had a known history of hereditary disease or cancer.
Table 2.
Clinical data of affected with familial spontaneous pneumothorax
| I−2 | II−2 | II−5 | II−8 | II−9 | II−11 | II−12 | II−14 | III−1 | III−3 | III−6 | III−7 | III−8 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Age (years) | 87 | 68 | 60 | 55 | 55 | 53 | 52 | 47 | 45 | 42 | 36 | 35 | 31 |
| Sex | F | F | M | F | F | F | F | F | F | M | M | F | F |
| Age at first attack of PSP | 70 | 68 | 46 | 31 | 50 | 48 | 34 | 38 | 36 | 27 | 22 | 29 | 24 |
| No. of PSP attack | 1 | 1 | 5 | 3 | 2 | 1 | 1 | 1 | 3 | 3 | 6 | 2 | 3 |
| PSP location | L | R | B | B | B | L | L | R | B | B | B | B | B |
| BMI (kg/m2) | 27.7 | 26 | 26.8 | 28.6 | 29.7 | 22.3 | 28.7 | 26.3 | 20 | 28.4 | 23.1 | 22.6 | 22 |
| Pulmonary bullae/blebs on CT scan | B&M | B&M | B&M | B&M | B&M | B&M | B&M | B&M | B&M | B&M | B&M | B&M | B&M |
| Treatment | TD | OBS | TD + Op | TD + Op | TD + Op | TD + Op | OBS | TD | TD + Op | TD + Op | TD + Op | TD | TD + Op |
Abbreviations: B&M, bilateral and multiple; B, bilateral; BMI, body mass index; CT, computed tomography; F, female; L, left; M, male; OBS, observation; Op, surgery operation; PSP, primary spontaneous pneumothorax; R, right; TD, tube drainage.
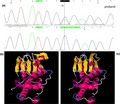
A novel heterozygous mutation c. 1273C>T (p.Gln425Ter) was detected in exon 11 of FLCN of the proband tested using Sanger sequencing. The novel mutation c. 1273C>T was also present in the family members (Figure 3). The deleterious annotation of genetic variants (DANN) analysis performed using VarSome Clinical revealed a DANN score of 0.9977 for the mutation we identified. This mutation results in a substitution of glutamine with terminal mutation which is a highly conserved residue among several species. Our analysis showed that conformational change in the mutated residue using Jmol 13.0 Software to compare the wild‐type and the mutant protein structure of FLCN (Figure 3).
Figure 3.
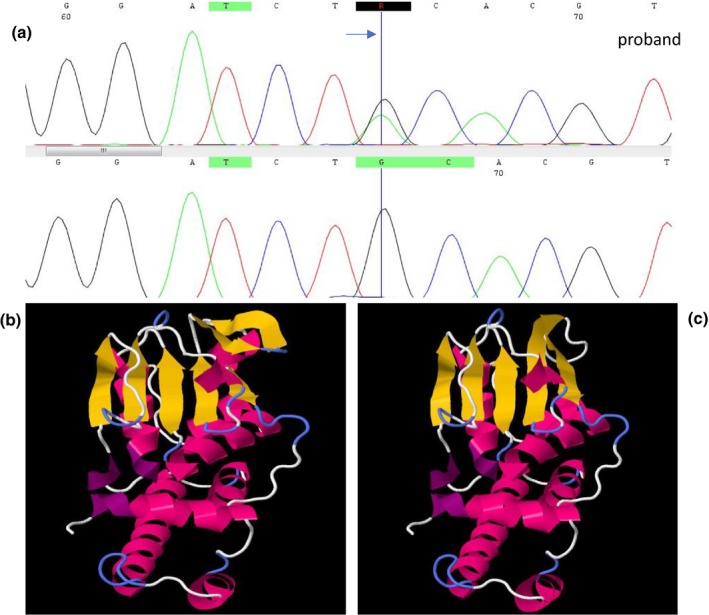
Chromatographs of sequence analysis of FLCN in proband and parents (a). A schematic diagram showing the wild‐type (b) and altered (c) protein structure in the residue where the mutation resides. FLCN, folliculin
4. DISCUSSION
In this study, we identified, for the first time, a nonsense mutation, which has not been previously reported in individuals diagnosed with PSP in the literature, in the FLCN of 13 members from three generations of the same family with a history isolated PSP.
To date, studies have reported the presence of more than 100 germline mutations in 14 exons of FLCN (Lim et al., 2010). These mutations in the FLCN have been identified in patients with BHD syndrome and patients with isolated familial PSP.
It has been reported that 50% of mutations in the FLCN are frameshift mutations resulting in a stop codon, while splice site, nonsense, missense, and deletion mutations are less prevalent (20%, 14.3%, 8.6%, and 4.3%, respectively) (Kim, Yoo, Kang, Cho, & Lee, 2012).
In this study, a heterozygous nonsense mutation was detected in exon 11 of FLCN of 13 members from three generations of the same family diagnosed with PSP. The c. 1273C>t mutation has not been reported as a variation in databases such as the Human Gene Mutation Database, 1000 Genomes, and Exome Variant Server. We believe that the single‐nucleotide mutation located in the c. 1273C>T (p.Gln425Ter) region of the FLCN result in a stop codon, thereby leading to protein truncation and losses of function. The DANN score ranges from 0 to 1 and a DANN score of 1 represents the highest possibility for pathogenicity. The DANN score of the identified mutation was found to be 0.9977.
Some mutations have been reported in different exons of the FLCN in cases of isolated familial spontaneous pneumothorax (Table 3). An inframe deletion mutation has been detected in exon 6 of FLCN in a Korean family with a history of recurrent PSP (Kim et al., 2012). A heterozygous C deletion (c.+ 1285) mutation in exon 11 has been found in the genetic analysis of an Indian family with a history of PSP (Ray, Paul, Chattopadhyay, Kundu, & Roy, 2015). A heterozygous mutation in exon 7 of FLCN was detected in a Swiss pedigree with a history of PSP (Fröhlich et al., 2008).
Table 3.
Reported mutations and their localizations in FLCN of familial spontaneous pneumothorax patients
| Gene | Mutations | Exon | Mutation type | Family phenotype | References |
|---|---|---|---|---|---|
| FLCN | c.1429C>T | 12 | Nonsense mutation | PSP | (Graham et al., 2005) |
| FLCN | c.943G>T | 9 | Nonsense mutation | PSP | (Graham et al., 2005) |
| FLCN | c.733delTCGG | 4 | Frameshift | PSP | Painter, Tapanainen, Somer, Tukiainen, & Aittomäki, 2005) |
| FLCN | c. 510C>G | 6 | Nonsense mutation | PSP | (Zhu, Shen, Zhu, & Tian, 2017) |
| FLCN | c.779G>A | 7 | Nonsense mutation | PSP | (Fröhlich et al., 2008) |
| FLCN | c.394G>A | 5 | Missense | PSP | (Fröhlich et al., 2008) |
| FLCN | c.924_926del | 6 | Inframe deletion | PSP | (Ren et al., 2008) |
| FLCN | c.1611_1631del | 10 | Frameshift | PSP | Ren et al., 2008) |
| FLCN | c.1740C>T | 11 | Missense | PSP | (Ren et al., 2008) |
| FLCN | c.1733insC | 11 | Frameshift | PSP | (Ren et al., 2008) |
| FLCN | c.1285delC | 11 | Frameshift | PSP | (Ray et al., 2015) |
| FLCN | nt1988 del GATG | 13 | Frameshift | PSP | (Gunji et al., 2007) |
| FLCN | nt1733 ins C | 11 | Frameshift | PSP | (Gunji et al., 2007) |
| FLCN | nt857 del C | 6 | Frameshift | PSP | (Gunji et al., 2007) |
| FLCN | nt852 ‐1 del gtccctccag | intron 5 | Inframe deletion | PSP | (Gunji et al., 2007) |
| FLCN | nt1795 ins CCACCCT | 12 | Frameshift | PSP | (Gunji et al., 2007) |
| FLCN | c.1537 del‐C | 10 | Deletion mutation | PSP | (Sundaram, Tasker, & Morrell, 2009) |
| FLCN | c. 1273C>T | 11 | Nonsense mutation | PSP | *Our mutation |
Abbreviation: FLCN, folliculin.
Mutations in exons 9 and 12 of FLCN have been reported to be associated with a higher number of cysts, larger cysts, and a higher incidence of pneumothorax (Toro et al., 2007). The frequency of renal neoplasm has been reported to be significantly lower in cytosine deletion mutations in exon 11 (Xing et al., 2017). Considering the previous studies, no definite distinction has been found between the type of FLCN mutation, BHD syndrome, and familial spontaneous pneumothorax. Therefore, it has been suggested that familial PSP cases should be evaluated in terms of BHD syndrome and the presence of fibrofolliculoma and renal mass should be excluded (Menko et al., 2009). In our study, the diagnosis of BHD syndrome was excluded because none of the patients had a history of fibrofolliculoma, renal cancer, or a different malignancy. FLCN mutations may also lead to different malignancies because it functions as a tumor suppressor gene. Possible genetic mutations that may cause isolated familial spontaneous pneumothorax are today still investigated. There is a need for further research on the subject.
In conclusion, we found that a heterozygous mutation in exon 11 of FLCN c. 1273C>T (p.Gln425Ter), which was identified for the first time in our study, might cause isolated familial spontaneous pneumothorax. The data suggest that autosomal dominant inheritance may be responsible for this transition. The presence of this mutation also confirms the familial inheritance of PSP in the proband.
We think that the causes underlying the molecular mechanism of familial spontaneous pneumothorax can be discovered through the examination of the connection between mutations in different exons of the FLCN and familial spontaneous pneumothorax.
CONFLICT Of INTEREST
None declared.
AUTHOR CONTRIBUTIONS
BGY and SC had the original idea for the study, BGY and EGT designed it. BGY sought institutional approval, compiled, checked and analyzed the data, managed the study database, computed the results, and wrote the initial draft of the manuscript including the text, tables, and figures. SSY and BGY together worked on the statistical analyses. BGY, EGT, SSY, and SC all critically reviewed the study design, contributed to the collection and analysis of the data and the interpretation of mutations and results, and commented on and approved the final manuscript. BGY submitted the study and is responsible as the guarantor for the overall content.
PATIENT CONSENT FOR PUBLICATION
The informed consent form was obtained from each family member involved in the study.
ETHICS APPROVAL
University of Health Sciences Haydarpasa Numune Training and Research Center (KAE).
ACKNOWLEDGMENT
This study was supported by the Scientific Research Project of the University of Health Sciences no. 2018/055.
Yavuz BG, Tanoglu EG, Salman Yılmaz S, Colak S. A novel FLCN mutation in family members diagnosed with primary spontaneous pneumothorax. Mol Genet Genomic Med. 2019;7:e1003 10.1002/mgg3.1003
Funding information
The authors did not apply for a specific grant for this research from any funding agency in the public, commercial, or nonprofit sectors. This study was supported by the Scientific Research Project of the University of Health Sciences no. 2018/055.
REFERENCES
- Abolnik, I. Z. , Lossos, I. S. , Zlotogora, J. , & Brauer, R. (1991). On the inheritance of primary spontaneous pneumothorax. American Journal of Medical Genetics, 40(2), 155–158. 10.1002/ajmg.1320400207 [DOI] [PubMed] [Google Scholar]
- Chiu, H. T. , & Garcia, C. K. (2006). Familial spontaneous pneumothorax. Current Opinion in Pulmonary Medicine, 12(4), 268–272. 10.1097/01.mcp.0000230630.73139.f0 [DOI] [PubMed] [Google Scholar]
- Delaney, J. C. , Gale, A. , & Walker, B. A. (1974). Familial spontaneous pneumothorax. Postgraduate Medical Journal, 50(588), 648–649. 10.1136/pgmj.50.588.648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engdahl, M. S. , & Gershan, W. M. (1998). Familial spontaneous pneumothorax in neonates. Pediatric Pulmonology, 25(6), 398–400. [DOI] [PubMed] [Google Scholar]
- Fröhlich, B. A. , Zeitz, C. , Mátyás, G. , Alkadhi, H. , Tuor, C. , Berger, W. , & Russi, E. W. (2008). Novel mutations in the folliculin gene associated with spontaneous pneumothorax. European Respiratory Journal, 32(5), 1316–1320. 10.1183/09031936.00132707 [DOI] [PubMed] [Google Scholar]
- Gibson, G. J. (1977). Familial pneumothoraces and bullae. Thorax, 32(1), 88–90. 10.1136/thx.32.1.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham, R. B. , Nolasco, M. , Peterlin, B. , & Garcia, C. K. (2005). Nonsense mutations in folliculin presenting as isolated familial spontaneous pneumothorax in adults. American Journal of Respiratory and Critical Care Medicine, 172(1), 39–44. 10.1164/rccm.200501-143OC [DOI] [PubMed] [Google Scholar]
- Gunji, Y. , Akiyoshi, T. , Sato, T. , Kurihara, M. , Tominaga, S. , Takahashi, K. , & Seyama, K. (2007). Mutations of the Birt Hogg Dube gene in patients with multiple lung cysts and recurrent pneumothorax. Journal of Medical Genetics, 44(9), 588–593. 10.1136/jmg.2007.049874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jmol: an open‐source Java viewer for chemical structures in 3D. Retrieved from http://www.jmol.org/
- Kim, J. , Yoo, J. H. , Kang, D. Y. , Cho, N. J. , & Lee, K. A. (2012). Novel in‐frame deletion mutation in FLCN gene in a Korean family with recurrent primary spontaneous pneumothorax. Gene, 499(2), 339–342. 10.1016/j.gene.2012.03.037 [DOI] [PubMed] [Google Scholar]
- Lenler‐Petersen, P. , Grunnet, N. , Jespersen, T. W. , & Jaeger, P. (1990). Familial spontaneous pneumothorax. European Respiratory Journal, 3(3), 342–345. [PubMed] [Google Scholar]
- Lim, D. H. , Rehal, P. K. , Nahorski, M. S. , Macdonald, F. , Claessens, T. , Van Geel, M. , … Maher, E. R. (2010). A new locus‐specific database (LSDB) for mutations in the folliculin (FLCN) gene. Human Mutation, 31(1), E1043–E1051. 10.1002/humu.21130 [DOI] [PubMed] [Google Scholar]
- Menko, F. H. , van Steensel, M. A. , Giraud, S. , Friis‐Hansen, L. , Richard, S. , Ungari, S. , … Maher, E. R. ; Consortium, E. B. (2009). Birt‐Hogg‐Dubé syndrome: Diagnosis and management. The Lancet Oncology, 10(12), 1199–1206. 10.1016/S1470-2045(09)70188-3 [DOI] [PubMed] [Google Scholar]
- Morrison, P. J. , Lowry, R. C. , & Nevin, N. C. (1998). Familial primary spontaneous pneumothorax consistent with true autosomal dominant inheritance. Thorax, 53(2), 151–152. 10.1136/thx.53.2.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter, J. N. , Tapanainen, H. , Somer, M. , Tukiainen, P. , & Aittomäki, K. (2005). A 4‐bp deletion in the Birt‐Hogg‐Dubé gene (FLCN) causes dominantly inherited spontaneous pneumothorax. American Journal of Human Genetics, 76(3), 522–527. 10.1086/428455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray, A. , Paul, S. , Chattopadhyay, E. , Kundu, S. , & Roy, B. (2015). Genetic analysis of familial spontaneous pneumothorax in an Indian family. Lung, 193(3), 433–438. 10.1007/s00408-015-9723-9 [DOI] [PubMed] [Google Scholar]
- Ren, H.‐Z. , Zhu, C.‐C. , Yang, C. , Chen, S.‐L. , Xie, J. , Hou, Y.‐Y. , … Yi, L. (2008). Mutation analysis of the FLCN gene in Chinese patients with sporadic and familial isolated primary spontaneous pneumothorax. Clinical Genetics, 74(2), 178–183. [DOI] [PubMed] [Google Scholar]
- Sahn, S. A. , & Heffner, J. E. (2000). Spontaneous pneumothorax. New England Journal of Medicine, 342(12), 868–874. 10.1056/NEJM200003233421207 [DOI] [PubMed] [Google Scholar]
- Sundaram, S. , Tasker, A. D. , & Morrell, N. W. (2009). Familial spontaneous pneumothorax and lung cysts due to a Folliculin exon 10 mutation. European Respiratory Journal, 33(6), 1510–1512. 10.1183/09031936.00062608 [DOI] [PubMed] [Google Scholar]
- Toro, J. R. , Pautler, S. E. , Stewart, L. , Glenn, G. M. , Weinreich, M. , Toure, O. , … Linehan, W. M. (2007). Lung cysts, spontaneous pneumothorax, and genetic associations in 89 families with Birt‐Hogg‐Dubé syndrome. American Journal of Respiratory and Critical Care Medicine, 175(10), 1044–1053. 10.1164/rccm.200610-1483OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Steensel, M. A. M. , Verstraeten, V. L. R. M. , Frank, J. , Kelleners‐Smeets, N. W. J. , Poblete‐Gutiérrez, P. , Marcus‐Soekarman, D. , … van Geel, M. (2007). Novel mutations in the BHD gene and absence of loss of heterozygosity in fibrofolliculomas of Birt‐Hogg‐Dubé patients. The Journal of Investigative Dermatology, 127(3), 588–593. 10.1038/sj.jid.5700592 [DOI] [PubMed] [Google Scholar]
- Wilson, W. G. , & Aylsworth, A. S. (1979). Familial spontaneous pneumothorax. Pediatrics, 64(2), 172–175. [PubMed] [Google Scholar]
- Xing, H. , Liu, Y. , Jiang, G. , Li, X. , Hou, Y. , Yang, F. , & Wang, J. (2017). Clinical and genetic study of a large Chinese family presented with familial spontaneous pneumothorax. Journal of Thoracic Disease, 9(7), 1967–1972. 10.21037/jtd.2017.06.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada, A. , Takeda, Y. , Hayashi, S. , & Shimizu, K. (2003). Familial spontaneous pneumothorax in three generations and its HLA. The Japanese Journal of Thoracic and Cardiovascular Surgery, 51(9), 456–458. 10.1007/BF02719603 [DOI] [PubMed] [Google Scholar]
- Zhu, J. F. , Shen, X. Q. , Zhu, F. , & Tian, L. (2017). Novel folliculin (FLCN) mutation and familial spontaneous pneumothorax. QJM: An International Journal of Medicine, 110(1), 23–26. 10.1093/qjmed/hcw109 [DOI] [PubMed] [Google Scholar]


