Figure 1.
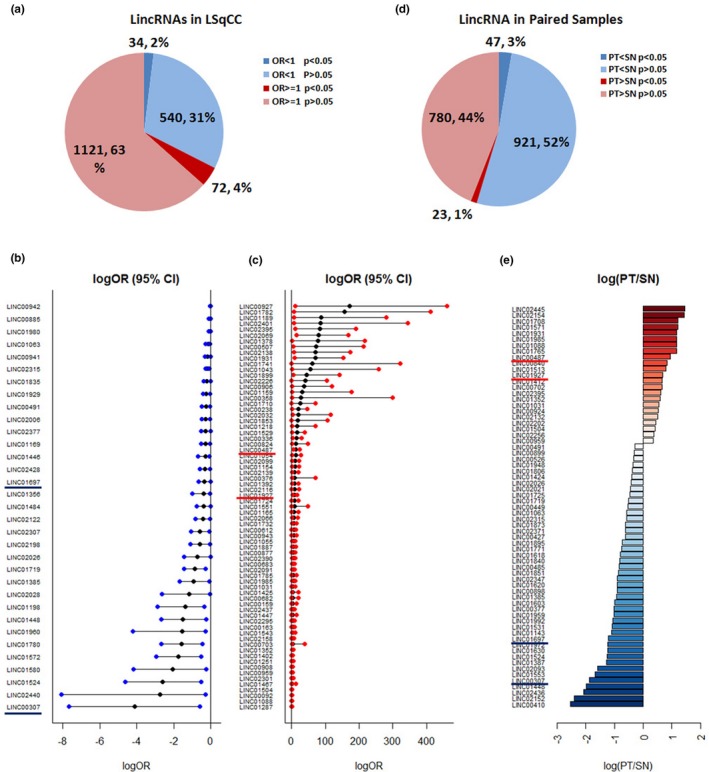
Transcriptional profile of lincRNAs in LSqCC tumor samples and solid normal tissue samples. (a) Transcriptional profile of 1,771 LincRNAs was compared between independent primary tumor LSqCC samples and solid normal tissue samples. 72 (4%) lincRNAs were significant risk predictive candidates for malignancies (OR > 1, p < .05), while 34 (2%) lincRNAs were siginificant protective candidates against malignancies (OR < 1, p < .05). (b) Natural logarithm of OR and 95% CI of OR, logOR (95% CI), of 34 significant protective lincRNAs. (c) Natural logarithm of OR and 95% CI of OR, logOR (95% CI), of 72 significant risk lincRNAs. (d) Transcriptional profile in paired primary LSqCC tumor samples and solid normal tissue samples. Gene expression level of 23 (1.30%) lincRNAs were significantly higher in primary LSqCC tumor samples than matched solid normal tissue samples (p < .05). The expression levels of 47 (2.65%) lincRNAs were significantly lower in primary tumors, compared to matched solid normal tissues (p < .05). (e). The natural logarithm value of the gene expression ratio of PT versus SN samples (log(PT/SN)) of significant lincRNAs that overexpressed (red) or lower‐expressed (blue) in primary LSqCC tumors, compared to their paired solid normal tissue samples
