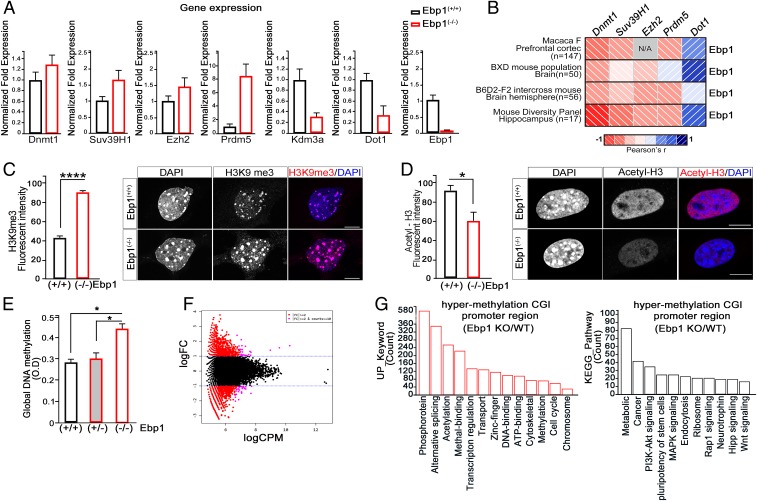
Fig. 3.
Ebp1 loss is associated with up-regulation of gene-silencing related genes. (A) Quantitative RT-PCR analysis of methylation related genes from E13.5 Ebp1(+/+) or Ebp1(−/−) yolk sacs. The relative fold changes were quantified and shown in the bar graphs. The mRNA levels were normalized to the levels of GAPDH. (B) Correlation matrices showing Pearson’s r between Ebp1 and the other genes (i.e., Dnmt1, Suv39h1, Ezh2, Prdm5, and Dot1) in the various species as indicated. See Dataset S1. (C and D) Fixed MEF cells (passage 2) were stained with anti-H3K9me3 (C) and anti-acetylation H3 antibody (D). The intensity of methylation and acetylation of H3 are shown as a bar graph. (Scale bar: 10 µm.) Data represent the mean ± SEM of 3 independent experiments. *P < 0.01, ****P < 0.0001 versus wild type. (E) gDNA was extracted from E13.5 embryonic brain. Global DNA methylation levels were measured and are shown as bar graphs. Data represent the mean ± SEM of 3 independent experiments. *P < 0.05. (F and G) MBD sequencing was performed in E15.5 Ebp1(+/+) and Ebp1(−/−) embryonic brain. (F) Smear plot of MBD-seq data showing the overall average (x axis) versus log2 fold change in methylation levels. Differentially methylated genes are shown in red, and nonsignificant changes are shown in black. (G) Hypermethylated CGI promoter in Ebp1(−/−)/Ebp1(+/+) mouse was subjected to GO-based classification using the DAVID gene-term classification tool. Annotation clusters with enrichment score >2.0 and P value <0.05 were selected as enriched functional categories.