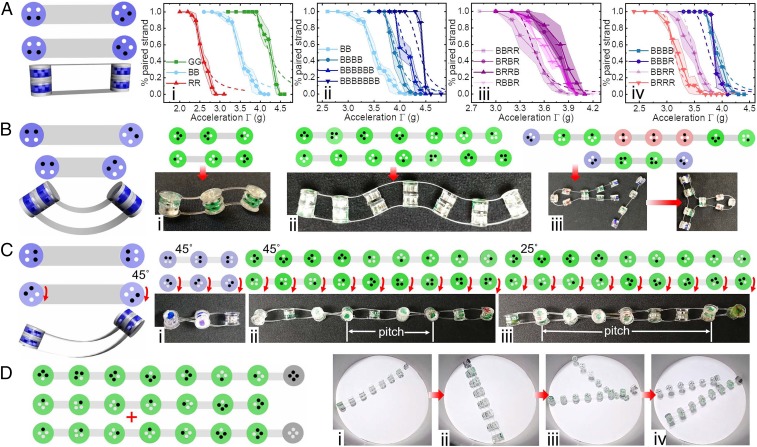
Fig. 3.
Magnetic strands and the resulting secondary structures. We make flat (A), bent (B), and twisted (C) structures from 2 complementary strands. (A) Percent of paired strands at different is sensitive to panel dipole pattern (i), number of pairing panels (ii), panel sequence (iii), and red-to-blue panel ratio (iv). The keys indicate the panel sequence and length. The shaded regions indicate the SE from 5 independent experimental measurements. The dashed lines are obtained from theoretical calculations. The broadening in the low probability region at high is expected since the experiment consists of measurements on a small number of paired strands (see SI Appendix for details). (B) Bending structures: shortest S-shaped strand (i), long S-shaped strand (ii), and hairpin structure (iii). (C) We create twisted structures by rotating the panels in 1 strand relative to the backbone (red arrows). (C, i) Shortest helix. (C, ii and iii) We control the pitch of the helices by changing the rotation angle of the panels from in ii to in iii. (D) Toehold exchange of a short complementary strand with a longer complementary strand. Here, spacers are used to weaken binding on 1 side of each strand. Initially, only the toehold region is exposed and available for strong binding. The leftmost panel shows a schematic of the initial pair of strands and the added longer strand (indicated by the ). (D, i–iv) Images from a movie of the toehold exchange. In snapshot D, i, we show the initial pair of strands. In snapshot D, ii, we show that the added longer complementary strand binds to both the toehold region and weakly to the other pair of strands. Such complexes do not have analogues in DNA. In D, iii, we observe that only the toehold region of the long chain, which consists of a single black panel, remains bound. Finally, in snapshot D, iv, the short strand is completely displaced by the long strand, completing the toehold exchange.