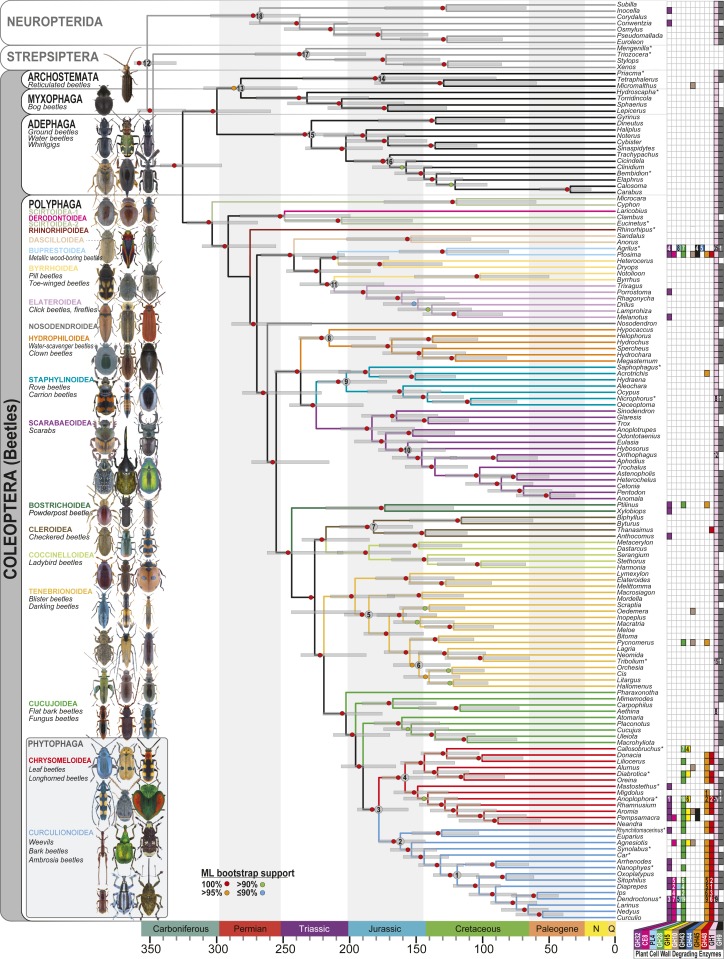
Fig. 1.
A dated phylogeny of beetles showing the distribution of putative PCWDEs, inferred from 4,818 nuclear genes (Methods and SI Appendix). Branches in suborder Polyphaga are color-coded by superfamily. Numbers indicate nodes constrained by fossil priors (SI Appendix, Table S5). Filled squares indicate the presence of putative PCWDEs and GH32 invertases (color-coded by gene family) based on analyses of whole-genome (asterisk) or RNA-Seq data. GH1 and GH9 have known ancient origins in metazoans (14, 47) and were expected to occur in most species. Asterisks denote results from the analysis of WGS (versus RNA-Seq) data. Numbers of homologs are indicated in each box when previously published (SI Appendix, Table S1). Note that Rhinorhipus was added after the initial analyses were completed based on a new ML tree search, which recovered the same topology. Bootstrapping was not conducted due to computational constraints. However, its placement was the same in the 521-taxon tree, where it had 100% ML bootstrap support. All higher taxa shown to illustrate morphological diversity (but not all species) were sampled. Cupes image courtesy of Matthew Bertone (North Carolina State University, Raleigh, NC). All other photos courtesy of Udo Schmidt (photographer).