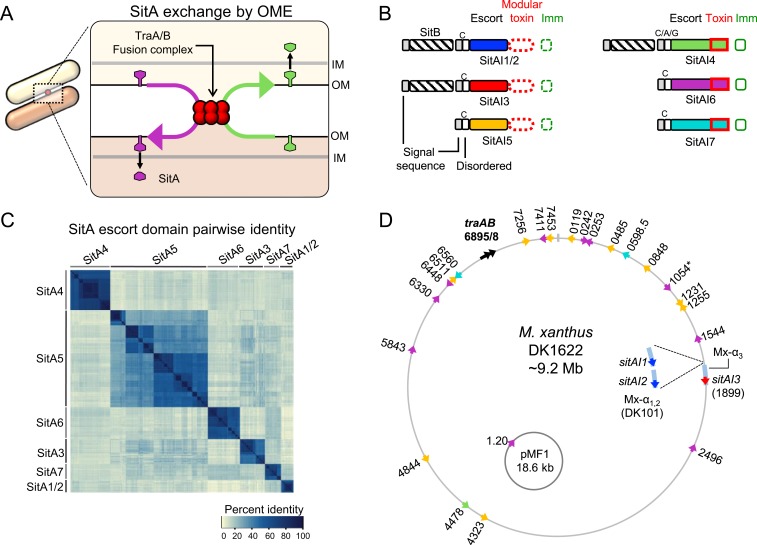
Fig. 1.
Four newly discovered SitA families. (A) Model for exchange of SitA by OME. Two adjacent cells with compatible TraA receptors form a TraAB OM fusion junction. Subsequently, transient membrane fusion allows the passive diffusion of SitA toxins between cells. SitA then traverses the cell envelope to access the cytoplasm, where they either act as nucleases or are inactivated by their allele-specific SitI immunity protein. (B) Overview of the domain organization of SitBAI and SitAI families. SitA families are classified into 2 groups by whether they have a modular toxic CTD (red dashed boxes) or a single polymorphic toxin domain (solid red boxes). Colors correspond to unique escort domains belonging to each family. Invariant lipobox cysteine (“C”) residues or alternative residues (in SitA4) are shown. Genes not to scale; Fig. 5 provides more detail. (C) Pairwise percent identity matrix of Myxococcaceae proteins from each SitA family. SitA proteins belonging to one family share homology with each other but not with proteins of a different SitA family. (D) Locations of sitA loci on the lab strain M. xanthus chromosome and the pMF1 plasmid from M. fulvus 124B02. DK101 is a parent strain of DK1622 that contains a ∼200-kb region of Mx-alpha that was spontaneously lost during the construction of DK1622, which harbors 2 additional sitA alleles (Inset). Numbers correspond to MXAN locus tags. Colors indicate SitA family based on the color scheme in B. Each sitA gene contains a cognate downstream sitI gene (not pictured). *MXAN_1054 has a frame-shift mutation.