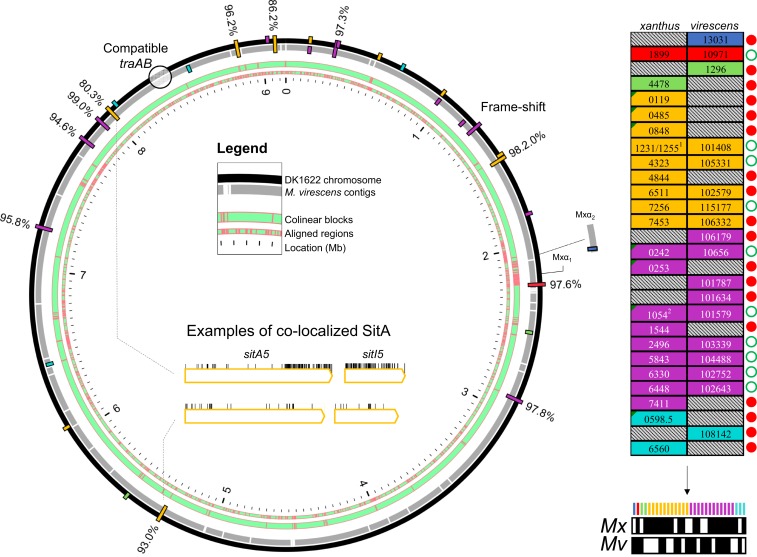
Fig. 6.
Comparative analysis of sitA genes from Myxococcus strains that are compatible for OME. (A) Whole-genome alignment depicts conserved and unique SitA toxins between 2 related Myxococcus genomes. Aligned regions are shown in green with red boundaries (“Legend”). Bars represent sitAI loci on the chromosomes. Bars spanning both genomes are colocalized sitA loci in regions of gene synteny. Inner-facing bars are unique M. virescens loci, whereas outer-facing bars are unique DK1622 loci. Amino acid percent identity between colocalized SitA alleles is shown. Inside the circular plot is an amino acid substitution analysis of one example of colocalized and conserved alleles (Lower) between the 2 genomes and colocalized alleles that have diverged at the SitA-CTD and SitI (Upper). Vertical lines indicate substitutions. (B) Total full-length sitA genes between the 2 genomes. MXAN and SAMN4488504 locus tag numbers are shown. Alleles that are predicted to be shared by both genomes (≥93% amino acid identity), i.e., reciprocal immunity, are placed side by side and marked with an open green circle. Genes unique to one genome are marked with a red circle. 1Although MXAN_1231 is an unmatched allele, its sitI is nearly identical to MXAN_1255 and so is not considered a unique allele, as M. virescens has a matching sitI downstream of SAMN4488504_101408. 2MXAN_1054 has acquired a frame-shift mutation, even though its SitI remains intact. The schematic (Bottom Right) is an overview of the total sitA loci from both genomes, whose presence in either genome is indicated by a black fill underneath the color-coded legend. Note that sitA1 and 2 are in the DK101 draft genome (Fig. 1), which is identical to DK1622 but contains 2 additional Mx-alpha regions that are not in the DK1622 genome and are not pictured here.