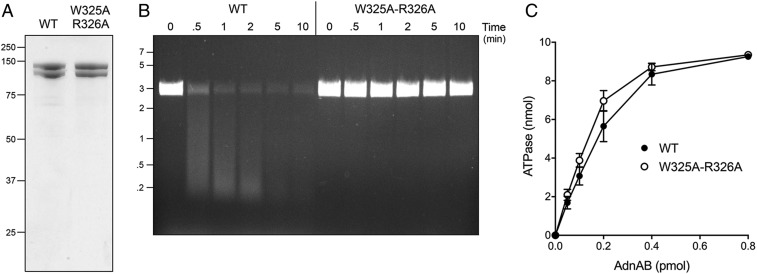
Fig. 5.
AdnB Trp325-Arg326 mutation uncouples ATP hydrolysis from DSB resection. (A) Aliquots (5 µg) of recombinant wild-type AdnAB and AdnA-AdnB(W325A-R326A) mutant heterodimers were analyzed by SDS/PAGE. The Coomassie blue-stained gel is shown. The positions and sizes (in kilodaltons) of marker polypeptides are indicated on the Left. (B) DSB resection. Reaction mixtures (50 μL) containing 20 mM Tris⋅HCl (pH 8.0), 2 mM MgCl2, 1 mM ATP, 200 ng linear pUC19 DNA (cut with SmaI), and 2.5 pmol of wild-type or mutant AdnAB were incubated at 37 °C. Aliquots (10 µL) were withdrawn at the times specified and the reactions were quenched immediately by adding 2.5 µL of 500 mM EDTA (pH 8.0). The reactions were supplemented with 5 μL of a solution containing 10 mM Tris⋅HCl (pH 7.6), 60% glycerol, 60 mM EDTA, and 0.15% Orange G and then analyzed by electrophoresis through a horizontal 0.8% agarose gel containing 45 mM Tris-borate, 1.2 mM EDTA, and 0.5 μg/mL ethidium bromide. DNA was visualized under shortwave UV illumination. (C) DNA-dependent ATPase activity. Reaction mixtures (10 µL) containing 20 mM Tris⋅HCl (pH 8.0), 2 mM MgCl2, 1 mM DTT, 1 mM (10 nmol) [α32P]ATP, 50 μM 85-mer ssDNA oligonucleotide, and AdnAB as specified were incubated for 10 min at 37 °C. The reactions were quenched by adding 2 μL of 5 M formic acid. Aliquots (2 μL) of the mixtures were applied to PEI-cellulose TLC plates, which were developed with 0.45 M ammonium sulfate. [α-32P]ATP and [α-32P]ADP were quantified by scanning the plates with a Fujix BAS2500 imager. The extent of ATP hydrolysis is plotted as function of input AdnAB. Each datum in the graph is the average of three independent titration experiments ± SEM.