Fig. 7.
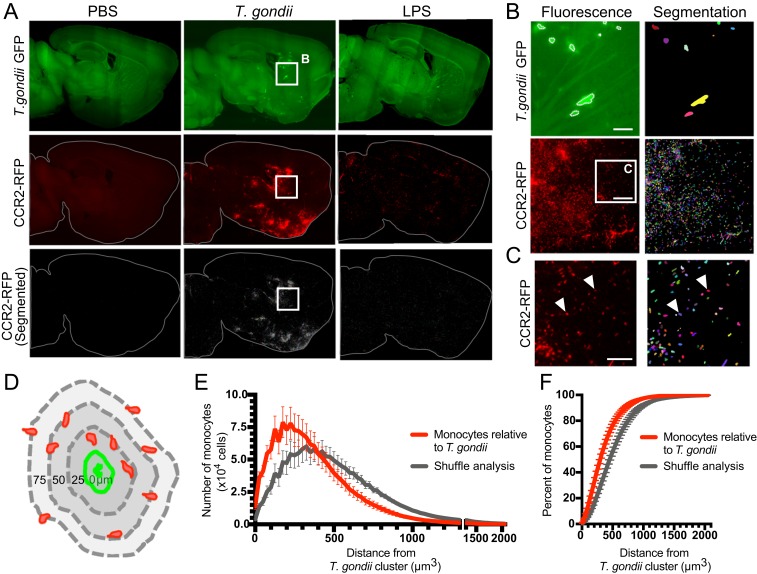
Monocytes are spatially correlated to T. gondii clusters. Brains from CCR2RFP/+ mice injected with PBS, T. gondii, or LPS were optically cleared using iDISCO+ and imaged by light-sheet microscopy. (A) Representative optical sections of GFP autofluorescence, GFP+ T. gondii, CCR2-RFP monocytes, and segmented (individually labeled) CCR2-RFP cells are shown. Contrast adjustment was matched between brains. (B) GFP+ T. gondii and CCR2-RFP cell segmentation show parasite clusters and individual monocytes (each denoted with a different color). (Scale bars, 100 µm.) (C) Magnified image of the CCR2-RFP panel in B, with white arrowheads indicating individual cells as identified by the segmentation algorithm. (Scale bars, 50 µm.) (D) Monocyte distance from each T. gondii cluster was measured in 3D space, as depicted. (E) Brain-wide analysis of monocyte distance from T. gondii clusters (red). Monocytes found within a T. gondii cluster have a distance of 0. Monocytes were randomly shuffled and distances of randomly distributed monocytes to T. gondii clusters were determined (gray). P < 10−7, Kolmogorov-Smirnov test. (F) The percent of monocytes at defined distances from T. gondii clusters is shown. Error bars represent SEM. nPBS = 5 mice, nT. gondii = 5 mice.