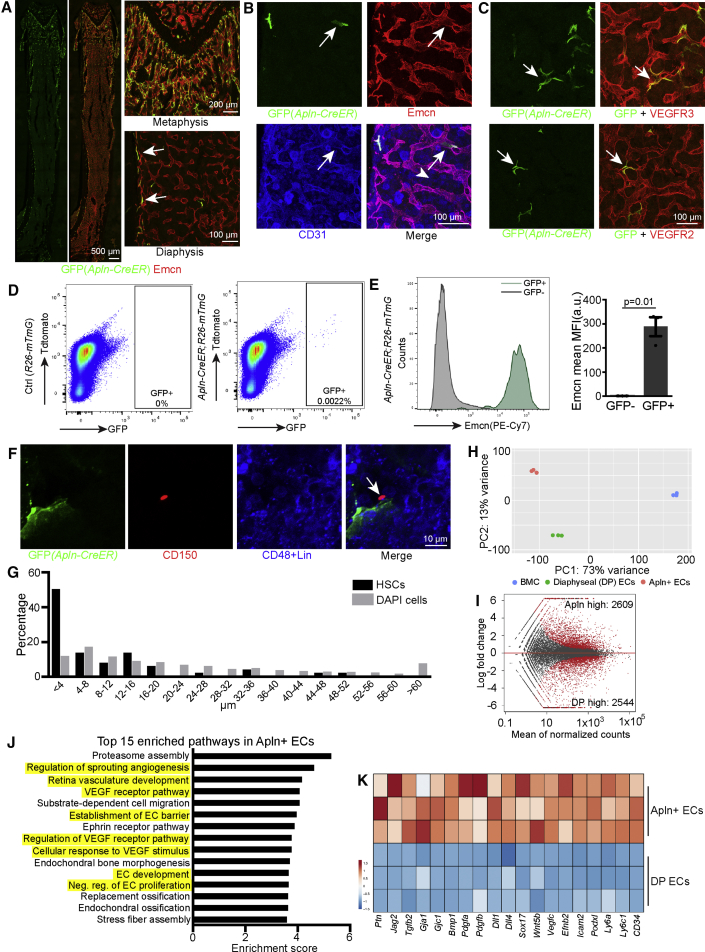
Figure 4.
Identification of Apln+ ECs in Adult BM
(A) Confocal tile scan images (left) showing distribution of GFP+ cells in Apln-mTmG mice. High magnifications (right) show enlarged regions of the metaphysis and diaphysis. Arrows mark GFP+ cells in endosteum.
(B and C) Emcn and CD31 (B) and VEGFR2 and VEGFR3 (C) staining in adult Apln-mTmG femur. Arrows mark GFP+ ECs.
(D) FACS plots of GFP+ cells isolated from Apln-mTmG mice.
(E) Histogram of Emcn expression in GFP– and GFP+ cells. Quantification of mean fluorescence intensity (MFI) in GFP– and GFP+ cells (n = 3 per group). Error bars, mean ± SEM. p values, two-tailed unpaired Students’ t test.
(F) CD150+ Lin– CD48– HSC (arrow) and GFP+ EC in the metaphysis and metaphysis-diaphysis transition area of Apln-mTmG mice.
(G) Distribution of distance between DAPI+ cells (n = 3,872) or CD150+ Lin– CD48– HSCs (n = 52) and GFP+ ECs cells in the metaphysis and metaphysis-diaphysis transition area of Apln-mTmG mice.
(H) PCA plot of total BM cells (BMC), Apln+ ECs (from whole bone), and diaphyseal ECs (DP ECs).
(I) MA plot showing differentially expressed genes between Apln+ ECs and DP ECs.
(J) Top 15 enriched pathways in GO analysis in Apln+ ECs relative to DP ECs.
(K) Heatmap of selected genes in Apln+ ECs.
See also Figure S4.