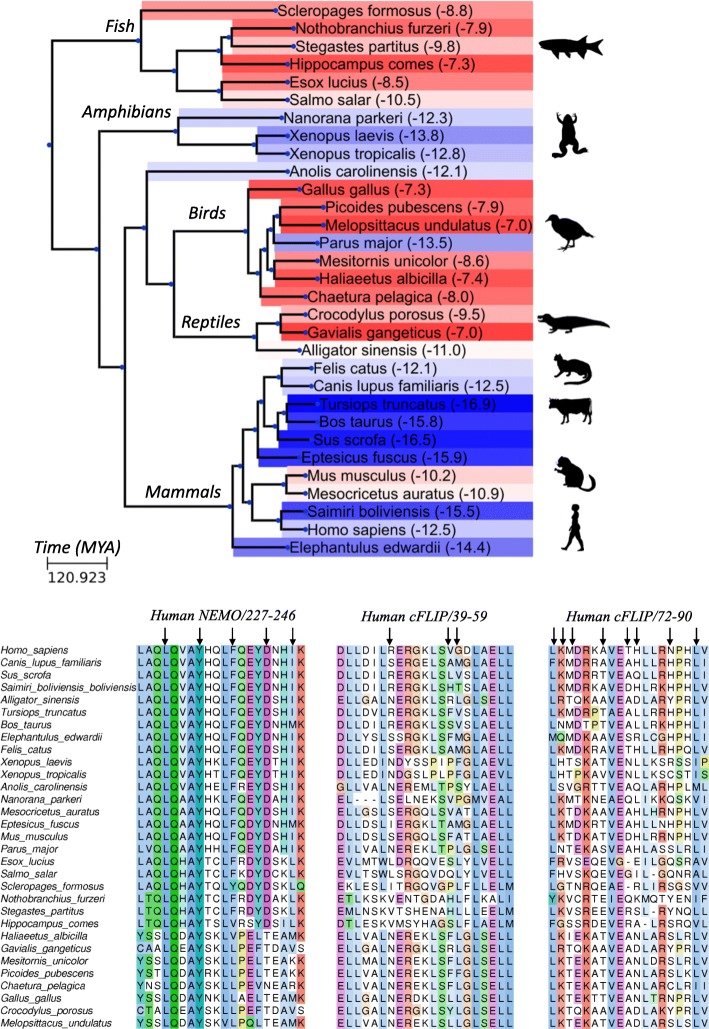
Fig. 7.
Analysis of evolutionary conservation of c-FLIP/NEMO binding sites. Phylogeny of the species and time points of the lineage diversification calculated using TimeTree database [23] are shown on the top of the figure. Background of the branches is colored based on predicted binding energy of c-FLIP and NEMO using FoldX. Reference organism names and predicted binding energies are indicated. Blue color gradient indicates an increase of stability of a complex in comparison with human complex; Red gradient indicates a decrease in stability. Silhouette images were obtained from PhyloPic (http://phylopic.org). The sequence alignments of NEMO and c-FLIP binding site regions are shown on the bottom of the figure. Arrows indicate amino acid residues located on the binding site interface of c-FLIP and NEMO