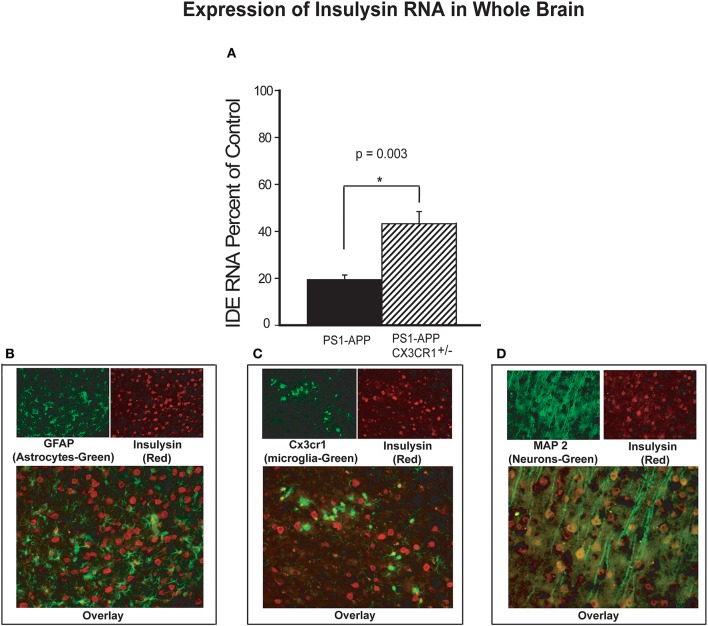
Figure 4.
Increased levels of insulysin RNA in brains of CX3CR1-deficient PS1-APP transgenic mice. (A) RNA was extracted from brains of PS1-APP, WT, PS1-APP-CX3CR1+/−, and CX3CR1+/− mice and assessed for expression of insulysin (IDE) by qPCR. Data represent mean percent of appropriate control ± SEM (n = 6 per genotype). (B–D) CX3CR1-dficient or WT mouse brain sections were co-stained for IDE and cell-specific markers and photographed by fluorescence microscopy at 40X magnification. (B) WT brain section co-stained for the astrocyte marker GFAP (green cells), for IDE (red cells). Most of the cells stained for IDE do not express GFAP. (C) CX3CR1-deficient brain section showing GFP-expressing microglia (green) and stained for IDE (red cells). Most of the microglia do not show detectable levels of IDE. (D) WT mouse brain slices co-stained for a neuronal marker MAP2 (green cells) and IDE (red cells). Most of the MAP2-expressing cells (neurons) also express IDE. * represent statistically significant.