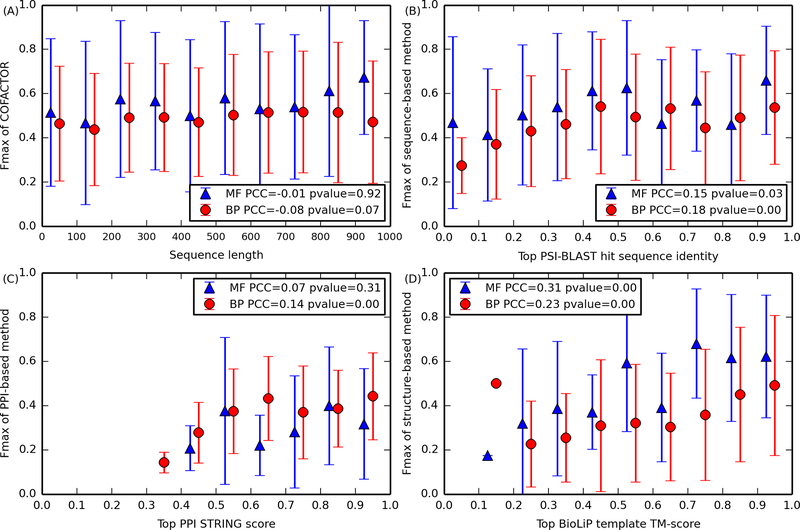
Figure 3.
Fmax of MF (blue triangle) and BP (red circle) GO term prediction versus (A) sequence length, (B) global sequence identity of closest PSI-BLAST hit, (C) highest PPI interaction score (STRING score), and (D) TM-score between query structure and the closest BioLiP template. A pair of error bars marks the standard deviation of Fmax at each bin. Inside each figure legend, the two numbers are the PCC and its p-value, respectively.33 (D) Among the set of 267 and 912 CAFA3 human targets for MF and BP, all were subjected to function prediction based on sequence and PPI by COFACTOR; only 88 and 227 targets, respectively, were predicted by the structure-based pipeline of COFACTOR.