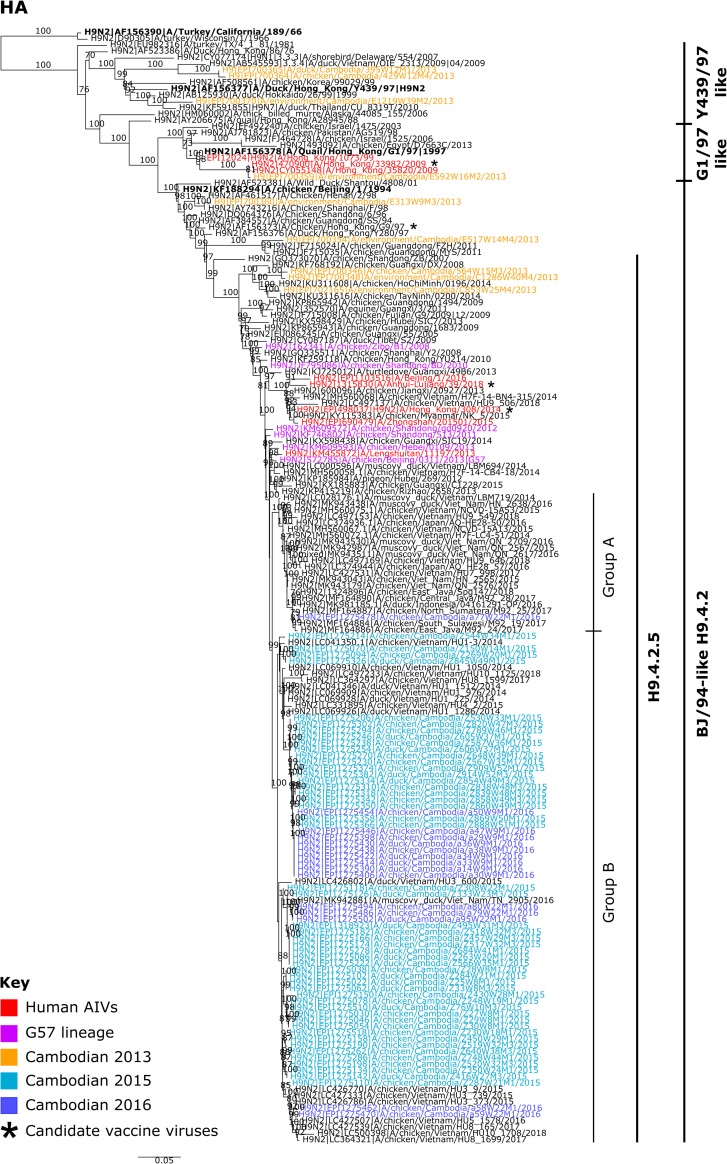
Fig 1. Maximum likelihood phylogenetic tree of the A(H9) HA gene.
The tree was produced using IQ-Tree with the GTR+ I + Γ model. Phylogenetic support was estimated using 1,000 ultrafast bootstrap replicates. Bootstrap values greater than 70% are displayed on branches. Cambodian A(H9) viruses are coloured according to the year they were detected: 2013 is shown in orange, 2015 in light blue and 2016 in dark blue. AIVs detected in humans are indicated in red and G57 viruses, as determined by Pu et al., 2015 [18], are shown in pink. Candidate vaccine viruses are indicated by an asterisks (*). Reference viruses are indicated in bold and viral lineages are indicated on the right of the phylogeny. Scale bars indicate the number of nucleotide substitutions per site.