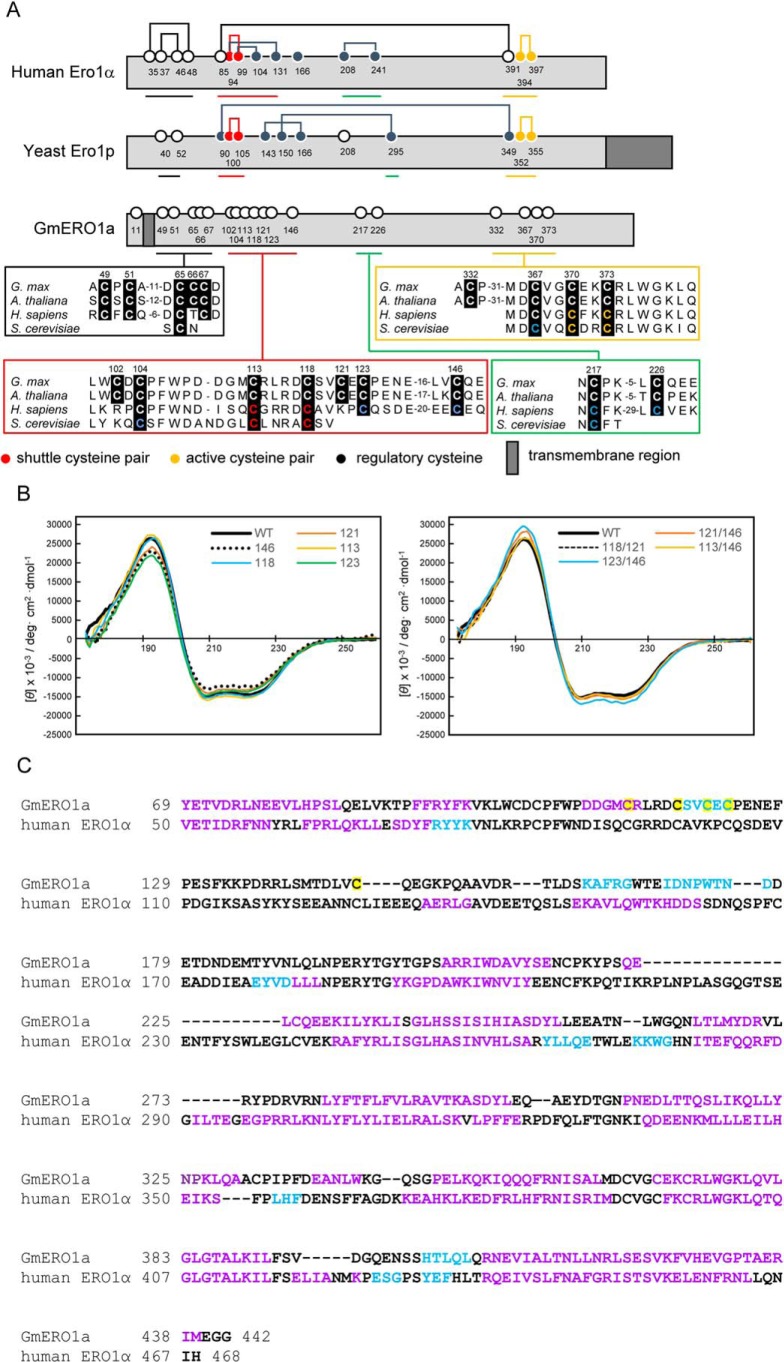
Figure 3.
Secondary structure of GmERO1a. A, schematic illustration and alignment of amino acid sequences around the cysteine residues of GmERO1a with human ERO1α and yeast ERO1p. Numbered circles indicate positions of cysteine residues. Lines connecting circles indicate disulfide bonds. B, VUVCD spectra of recombinant wildtype (WT) GmERO1a and mutants in which cysteine residues were substituted with alanine residues. Colored lines in each spectrum indicate mutants in which the indicated cysteine residues were replaced with alanine residues. The number on each spectrum indicates the cysteine residue replaced with alanine. C, alignment of secondary structures of GmERO1a and human ERO1α. Secondary structures of GmERO1a were predicted from the VUVCD–NN method. Secondary structural data of human ERO1α were obtained from the crystal structure (Protein Data Bank code 3AHR). α-Helix, β-strand, and coil regions are indicated in purple, light blue, and black in the amino acid sequences, respectively. Cys-113, Cys-118, Cys-121, Cys-123, and Cys-146 are highlighted in yellow.