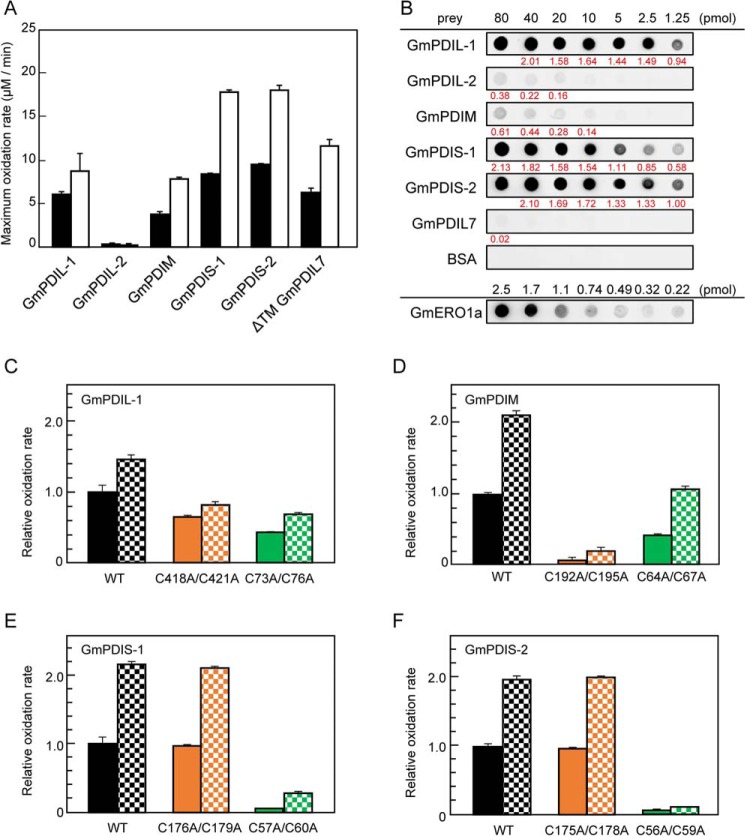
Figure 6.
Substrate specificity of GmERO1a for PDI family proteins. A, oxidation of each PDI family protein by WT GmERO1a (black bars) or C121A/C146A-hyperactive mutant GmERO1a (HA) (white bars), determined as described in the legend for Fig. 4B. B, GmERO1a binds to PDI family proteins with different affinities. Far-Western blot analysis was performed with each PDI family protein as prey and GmERO1a as bait. Bound GmERO1a was detected using anti-GmERO1a serum. At the bottom lane (GmERO1a), 2.5, 1.7, 1.1, 0.74, 0.49, 0.32, or 0.22 pmol GmERO1a was dotted as a positive control. The amount of GmERO1a bound on each spot of PDI family proteins was calculated based on the dot intensities and GmERO1a standard dots using ImageJ software. All experiments were performed on the same nitrocellulose membrane. Value written in red below each dot indicates the amount (picomoles) of GmERO1a bound on each dot. C–F, relative ratio of maximum oxidation rates WT GmPDIL-1 (C), GmPDM (D), GmPDIS-1 (E), and GmPDIS-2 (F) and respective active-cysteine residue mutants by WT (solid bars) or HA GmERO1a (checkered bars). Numbers in the designations of mutants indicate cysteine residues substituted with alanine residues. The rates of oxidation of PDI family proteins were monitored as amount of NADPH consumed in the presence of 1 μm WT or HA GmERO1a and 3 μm of each PDI family protein or respective mutant in the presence of 3 mm GSH, 120 μm NADPH, and 1 unit/ml GSH reductase. Data are presented as the mean ± S.E. of n = 3 experiments.