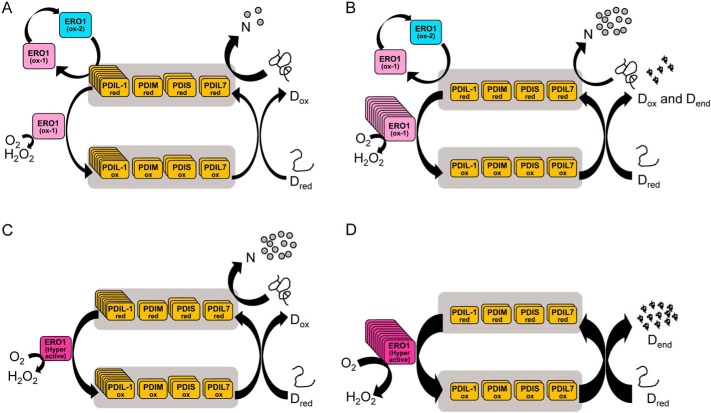
Figure 8.
Model of oxidative protein folding by the multiple PDI family proteins/ERO1 system in plants. A, oxidative protein folding at a proper molar ratio of WT ERO1 to PDI family proteins. Oxidized PDI family proteins introduce disulfide bonds in unfolded protein (Dred) producing denatured protein with nonnative disulfide bonds (Dox), and reduced PDI family proteins rearrange the disulfide bonds to the native ones, producing correctly folded native protein (N). Reduced PDI family proteins convert inactive ERO1 (ox-2) to the active form (ox-1). B, oxidative protein folding at an improperly high molar ratio of WT ERO1 to PDI family proteins. Oxidized PDI family proteins introduce disulfide bonds in Dred, and reduced PDI family proteins produce N. However, too many disulfide bonds beyond isomerization capacity of PDI family proteins are supplied by ERO1, resulting in part of Dred becoming denatured proteins with dead-end disulfides (Dend). C, oxidative protein folding at a proper molar ratio of hyperactive ERO1 to PDI family proteins. Many disulfide bonds within the limit of isomerization capacity of PDI family proteins are supplied by hyperactive ERO1 to Dred, resulting in Dred becoming N effectively. D, oxidative protein folding at an improperly high molar ratio of hyperactive ERO1 to PDI family proteins. Too many disulfide bonds are supplied by hyperactive ERO1, resulting in most of Dred becoming Dend. The number of copies of boxes are reflecting the relative concentration of the proteins.