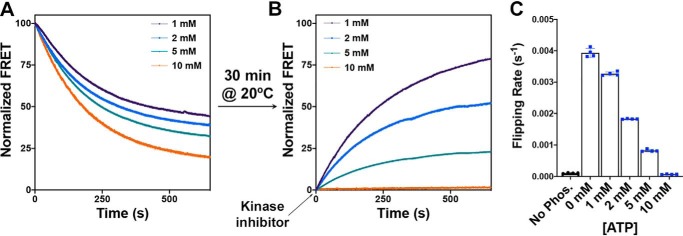
Figure 5.
Determination of the rate of dephosphorylation-induced topological switch of a membrane protein after inhibition of kinase activity. A and B, real-time FRET measurements monitoring phosphorylation-induced topological switch of EMD C6 in the LacY −2/0/−2 template reconstituted in proteoliposomes made of E. coli total lipids before (A) and after (B) addition of kinase inhibitor. LacY proteoliposomes with encapsulated phosphatase were incubated at 20 °C for 30 min after addition of excess PDK1. When a fluorescence steady state was reached, the PDK1 inhibitor GSK 2334470 was added. The various traces depict the topological switch of EMD C6 with various amounts of ATP encapsulated in the proteoliposomes, allowing the determination of kF2. Data represent the averaged normalized fluorescence expressed as the ratio FMeas/F0. C, the flipping rates, kF2, determined from single exponential fit of the data shown in B. In all cases, the data represent mean values ± S.D. from four experimental replicates.