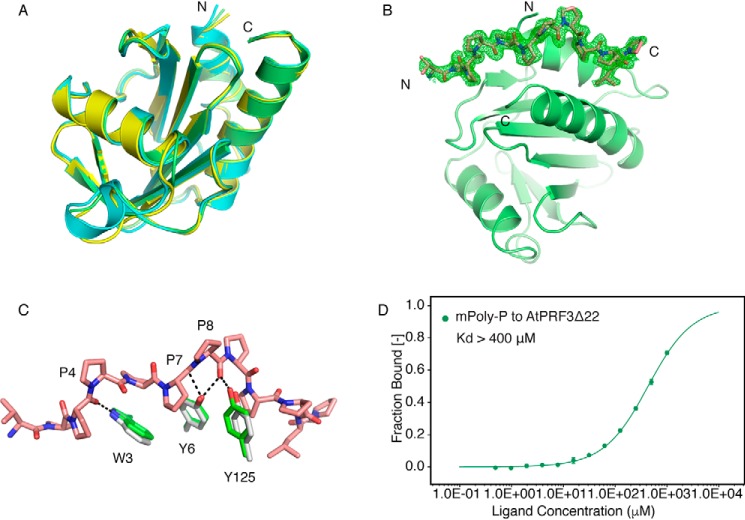
Figure 6.
Structural comparison of AtPRFs and the Poly-P binding. A, crystal structure of AtPRF1 (yellow) (PDB code 3NUL) and AtPRF3Δ37 (cyan) were aligned to AtPRF2 (lime). B, unbiased omit Fo − Fc electron density map (green) contoured at 2.2σ of Poly-P (salmon) bound AtPRF2 (lime). C, the alignment of AtPRF2 to Poly-P–bound AtPRF2. Whereas the Apo structure is shown in gray, the complex structure is colored in green, and the Poly-P is shown in salmon. The residues involved in interaction are labeled, respectively. D, microscale thermophoresis binding curves of the mPoly-P titration curve to AtPRF3Δ22 with the dissociation constant indicated. All the measurements were performed with three biological replicates.