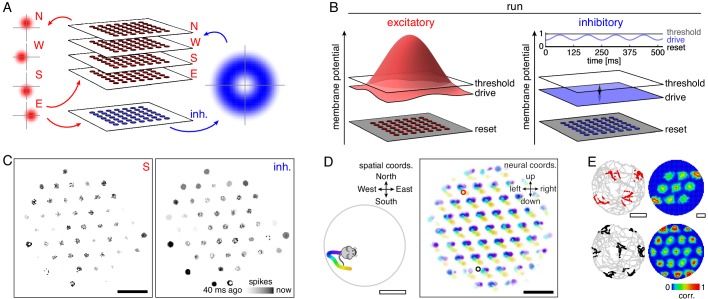
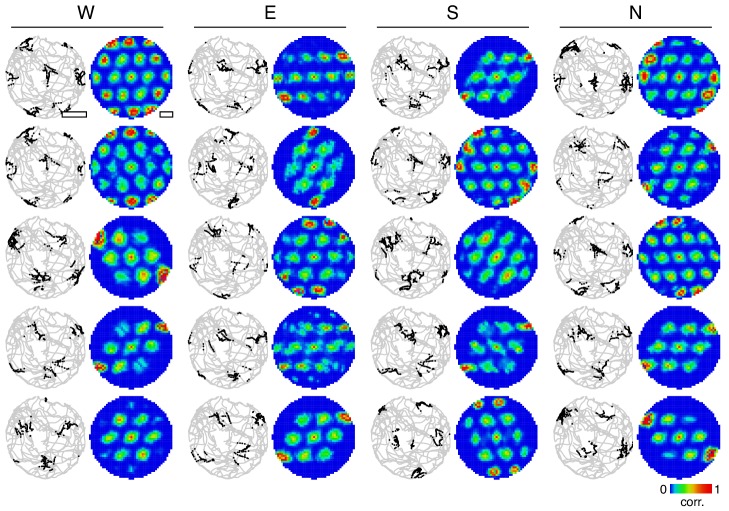
Figure 1. Model architecture and generation of 2D grid cells.
(A) Our model consists of a neural sheet with five overlapping populations, four of them excitatory—N, W, S, and E—and one inhibitory. Each density plot depicts the synaptic outputs in the sheet of a neuron at the origin. (B) Each neuron is driven to a particular membrane potential, which exceeds the spiking threshold for excitatory neurons at the center of the sheet and oscillates at 8 Hz for inhibitory neurons while the animal is running. (C) Snapshot of neural activity showing S and inhibitory populations separately; other excitatory populations have activity patterns similar to that of the S population. Each pixel is a neuron and dark colors indicate recent spiking. (D) Left, segment of a 2D open field trajectory. Right, neural activity over the course of the segment with each neuron colored according to the position at which it attained its maximum firing rate. Each attractor bump moves in synchrony with animal motion. (E) Left, two sample grid cells with spikes shown as colored dots superimposed on the animal’s trajectory. Each neuron’s location in the sheet is indicated by a circle of corresponding color in D. Right, autocorrelation of rate maps calculated from spikes at left. Black scale bars, 50 neurons. White scale bars, 50 cm.