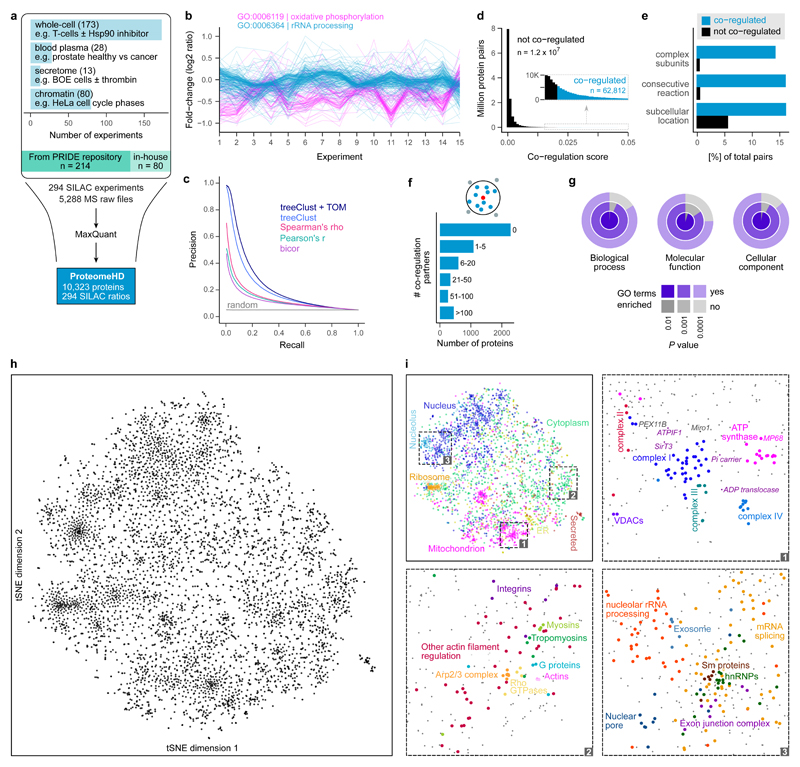
Figure 1. Co-regulation map shows associations between human proteins.
(a) Assembly of ProteomeHD, which quantifies the protein response to 294 perturbations using SILAC 34. Most measurements document protein abundance changes in whole-cell samples, but in some cases subcellular fractions were enriched to detect low-abundance proteins. Data were collected from PRIDE 35 and produced in-house. (b) A random set of experiments from ProteomeHD, showing that groups of proteins with related functions, e.g. Gene Ontology 41 (GO) biological processes, display similar expression changes. Note that the fold-changes are often very small. (c) Precision - recall analysis showing that the treeClust 37,38 algorithm outperforms three correlation-based coexpression measures. Applying the topological overlap measure (TOM) improves performance further. Annotations in Reactome 36 were used as gold standard. (d) Co-regulation scores for all protein pairs are obtained by combining treeClust with TOM. The score distribution is highly skewed. Where an arbitrary threshold is required, the highest-scoring 0.5% of pairs (N = 62,812) are considered “co-regulated”. (e) Co-regulated protein pairs are strongly enriched for subunits of the same protein complex, enzymes catalysing consecutive metabolic reactions and proteins with identical subcellular localization. (f) Most proteins are co-regulated with no or few other proteins, but many have more than 5 co-regulated partners. (g) Considering proteins that are co-regulated with ≥10 proteins, these groups of co-regulated proteins are almost always enriched in one or more GO terms. (h) The global co-regulation map of ProteomeHD created using t-Distributed Stochastic Neighbor Embedding (t-SNE). Distances between proteins indicate how similar their expression patterns are. See www.proteomeHD.net for an interactive version of the map. n = 5,013 proteins. (i) The co-regulation map broadly corresponds to subcellular compartments, and more detailed functional associations can be observed at higher resolution, as exemplified in subpanels 1-3.