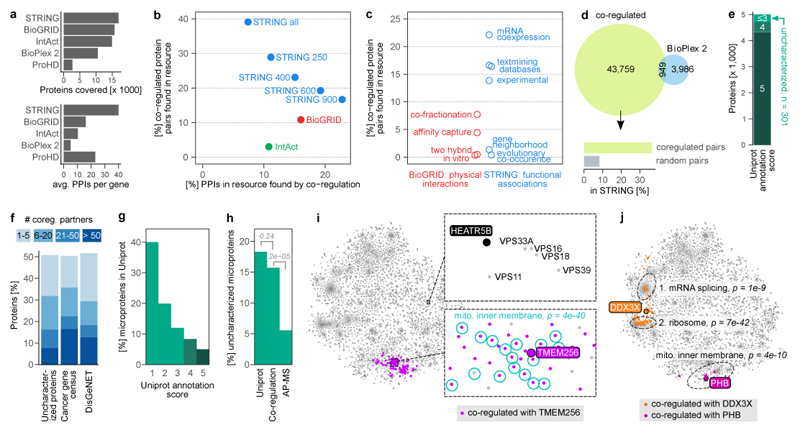
Figure 3. Protein co-regulation predicts functions of unknown proteins.
(a) Coverage of protein - protein interactions (PPIs) in comparison to other resources. Top barchart shows the number of genes covered, i.e. having at least one PPI above cut-off. STRING cut-off used: medium (400). Bottom chart shows the average number of PPIs of covered genes. The co-regulation map (ProHD) covers fewer genes than STRING, BioGRID, IntAct and BioPlex 2, but covers many associations between those genes. (b) Overlap between PPIs discovered by protein co-regulation and PPIs already present in large-scale annotation resources that cover both physical (BioGrid and IntAct) and functional (STRING 69) associations. Multiple association score cut-offs were considered for STRING. These three resources integrate data from many small and large-scale studies. (c) Coverage of co-regulated protein pairs in BioGRID and STRING broken down by the type of functional genomics evidence available in each resource. (d) Number of co-regulation links compared to PPIs found for the same set of genes by BioPlex 2.0 2, one of the largest PPI datasets reported to date by a single study. Associations unique to co-regulation are strongly enriched for links in STRING, compared to random gene pairs. (e) Out of the 5,013 proteins in the co-regulation map, 301 have a UniProt annotation score ≤3 and are thus defined as uncharacterized. (f) Connectivity of either uncharacterized proteins or proteins encoded by disease genes to well-characterized proteins (annotation score ≥4). 51% of uncharacterized proteins have at least one co-regulation partner, 32% have more than five. (g) Barchart showing the percentage of all 20,408 human UniProt (SwissProt) proteins that are microproteins, i.e. have a molecular weight < 15 kDa. Note that microproteins are heavily enriched among less well-characterized proteins. (h) 18% of 5,187 uncharacterized proteins in UniProt are microproteins, compared to 16% of the 153 uncharacterized proteins in the co-regulation map and 6% of 1,422 uncharacterized proteins in state-of-the-art AP-MS experiments, represented by BioPlex. P-values are from one-sided Fisher’s Exact test. (i) The uncharacterized microprotein TMEM256 has many co-regulation partners (n = 130), which are enriched for GO term “mitochondrial inner membrane” (n = 42) among others. Bonferroni-adjusted P-value is from a hypergeometric test. The uncharacterized HEATR5B protein has no co-regulation partners above the default threshold, but its position in the map nevertheless indicates a potential function. (j) For multifunctional proteins, co-regulation can reveal a mix of their functions (DDX3X; n = 14 of 81 co-regulated proteins annotated with GO term “mRNA splicing, via spliceosome, n = 27 with GO term “cytosolic ribosome”), or their main function only (prohibitin, PHB; n = 9 of 11 co-regulated proteins annotated as “mitochondrial inner membrane”). Three representative GO terms are shown.