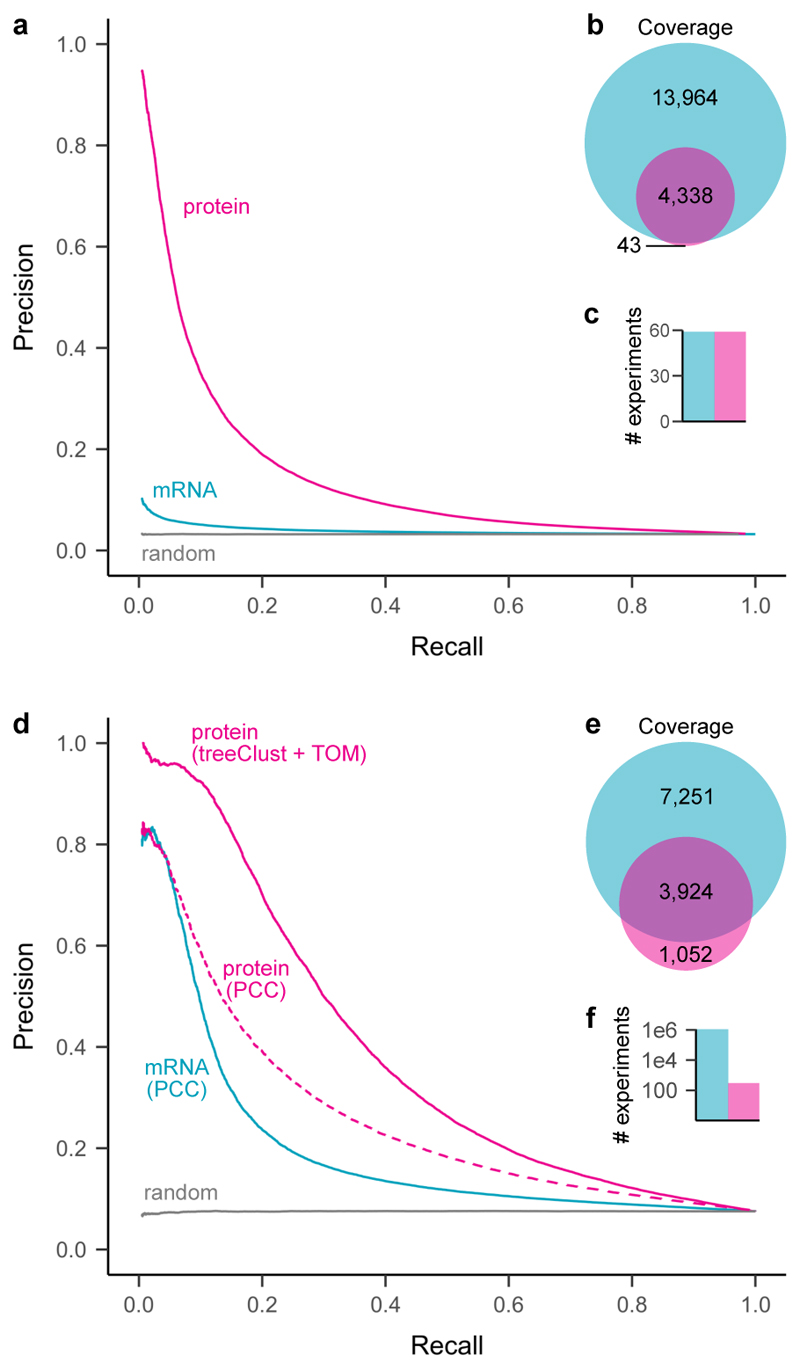
Figure 5. Protein co-regulation enables higher precision but lower coverage than mRNA coexpression.
(a) Precision-recall analysis of treeClust machine-learning on a subset of ProteomeHD, that is 59 samples for which matching RNA-seq data were available from a separate study 67.
Reactome pathways were used as gold standard for true functional associations (proteins found in same pathway) and false associations (never found in same pathway). Only annotated genes covered by both datasets were considered for PR analysis (n = 2,901). (b) Venn diagram showing number of genes covered by each analysis. (c) Barchart showing number of experiments the curves are based on. (d) Similar precision-recall analysis of treeClust machine-learning on the full ProteomeHD database, in comparison to Pearson correlation obtained by STRING 69 on the basis of one million human mRNA profiling samples deposited in the NCBI Gene Expression Omnibus 68 ("mRNA / PCC"). Protein co-regulation outperforms mRNA correlation despite being based on orders-of-magnitude less data. This is partially due to the use of machine-learning, as predicting associations from ProteomeHD using PCC decreases performance markably ("protein / PCC"). Only annotated genes covered by both datasets were considered for the PR analysis (n = 2,743). (e, f) same as (b, c).