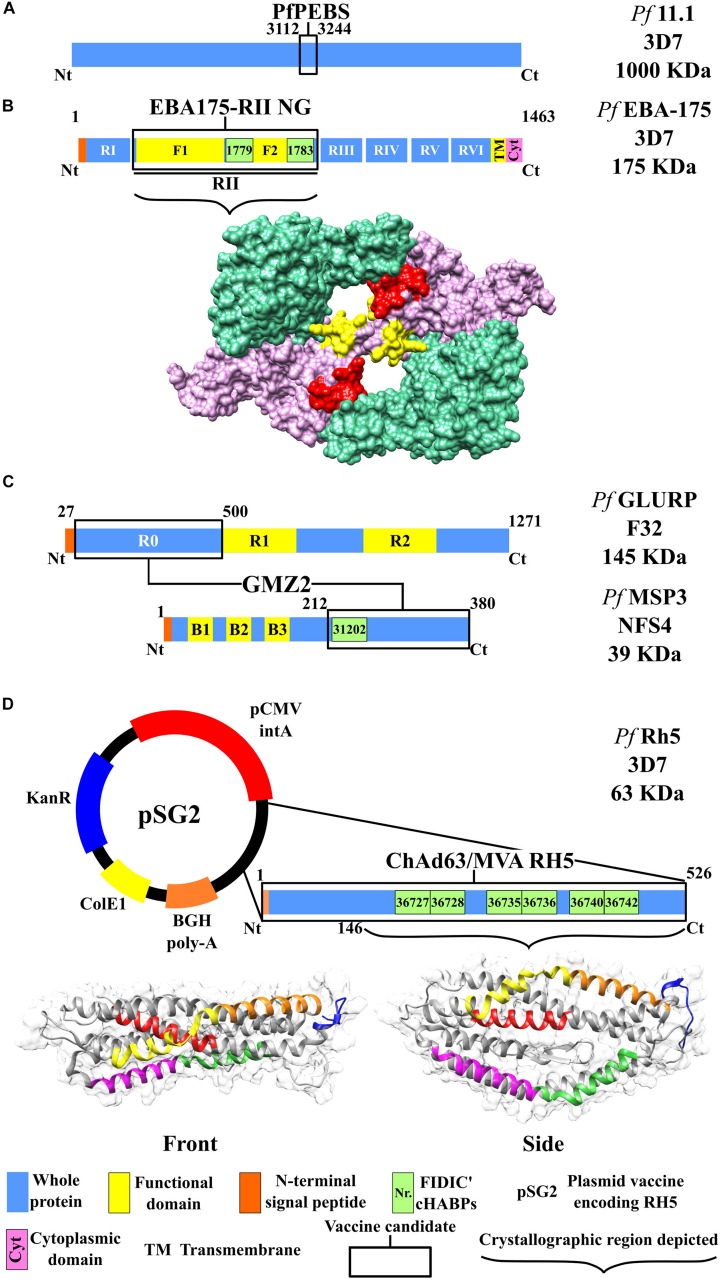
FIGURE 3.
Scheme for the proteins used as candidates for an antimalarial vaccine (II). Color code according to that used in Figure 2. (A) PfPEBS vaccine. (B) Top: Scheme for the EBA-175 RII NG vaccine. Bottom: Protein structure, including region II surface, F1 (green) and F2 domains (pink), and conserved high-activity binding peptides (cHABPs) 1779 (red) and 1783 (yellow), PDB 1ZRO (Tolia et al., 2005). (C) GMZ2 vaccine, including the scheme for proteins MSP3 and GLURP according to the color code used for Figure 2. (D) ChAd63/MVA RH5 vaccine. Top: pSG2 plasmid, including the kanamycin resistance gene (KanR), cytomegalovirus (CMV) with the associated intron A (CMVintA), bovine growth hormone (bGH) with polyadenylation (bGH PolyA), Escherichia coli β-galactosidase (ColE1), and the RH5-encoding gene. Bottom: Surface and ribbon for the RH5 protein with cHABPs 36727 (magenta), 36728 (green), 36735 (yellow), 36736 (orange), 36740 (red), and 36742 (blue). PDB 4WAT (Chen et al., 2014).