Figure 3.
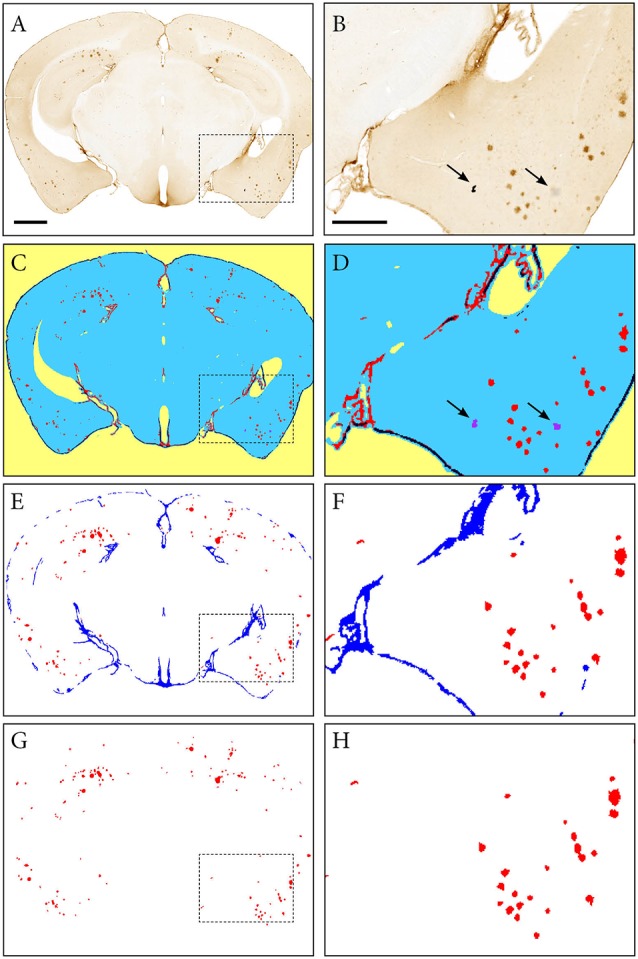
Segmentation of images with the ilastik software. An image of a Tg2576 mouse brain section, immunohistochemically labelled for pan-Aβ (4G8), processed with the pixel and object classification workflows in the ilastik software (version 1.2.2. post2). Panels (A,C,E,G) show the whole image, with (B,D,F,H) representing the area identified in the dashed box. (C,D) show the output of the pixel classification workflow, with images segmented into five classes based on differences in intensity, edge and texture (red: specific immunohistochemical labelling, blue: unlabelled tissue, purple: artefacts, black: non-specific labelling, yellow: background). The pixel classification workflow is able to differentiate labelling and artefacts such as marks on the coverslip and debris (see arrows). Panels (E,F) show the output of the object classification workflow: the probability maps derived from the pixel classification workflow were thresholded at 0.4 for the channel representing the labelling, and classified into two classes based on object-level features such as size and shape (red: β-amyloid plaques, blue: non-specific labelling). Panels (G,H) show the object classification output with the blue channel removed to visualize the β-amyloid plaques only. Images (A,C,E,G) are displayed at the same magnification with the scale bar representing 1 mm. The scale bar for figures (B,D,F,H) represents 500 μm.
