Figure 2.
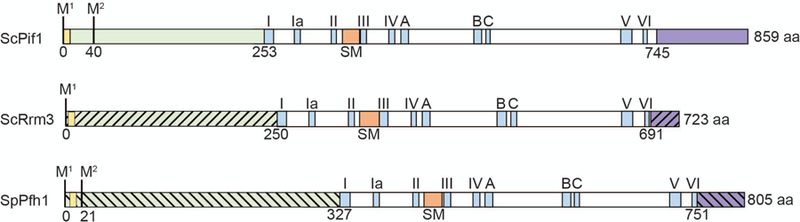
Schematics of the structures of three Pif1 family DNA helicases: S. cerevisiae Pif1, S. cerevisiae Rrm3, and S. pombe Pfh1. Each protein contains an amino-terminal domain (green) and a carboxyl-terminal domain (purple): the slashes indicate that the amino and carboxyl termini are divergent in sequence and size among the three helicases. The amino-terminal domain contains a putative mitochondrial targeting sequence (yellow) that has been confirmed for ScPif1 and SpPfh1. The first and second methionine marking the start of the mitochondrial (M1) and nuclear (M2) isoforms are indicated. The positions of the seven conserved helicase motifs are indicated by blue rectangles and roman numerals. The Pif1 signature motif (SM) distinguishes Pif1 family helicases from other SFI helicases and like the seven canonical helicase motifs is essential for ATPase activity (orange rectangle). Three motifs of unknown function that have high homology to E.coli RecD are indicated (blue rectangles; labeled A, B and C). The helicase domains (in white) are ~40% identical in all pair-wise combinations. The boundaries of the amino-terminal domains are based on the first amino-acid in helicase motif I; the boundaries of the carboxyl terminal domain are determined by the last amino acid in helicase motif VI.
