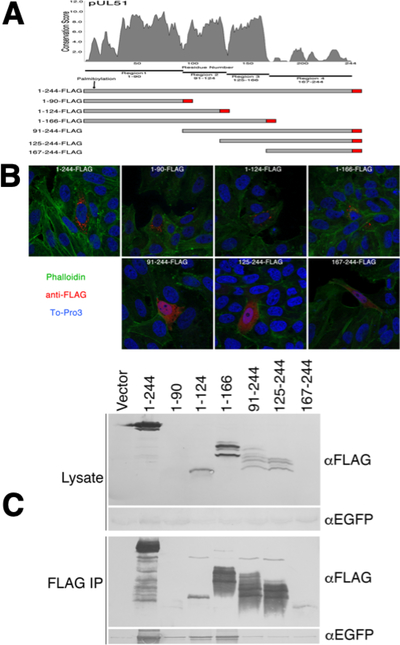
Figure 1.
Localization and pUL7 interactions of pUL51 truncations. (A) Conservation plot of pUL51 protein coding sequence and schematic diagram of FLAG-tagged pUL51 truncations. The plot shows conservation of biochemical properties of amino acids using all available herpesvirus pUL51 homologous sequences aligned using the program MUSCLE (61). Each residue position receives a conservation score, and scores were averaged over a sliding 5 amino acid residue window. The schematic underneath the plot shows the boundaries of the pUL51 truncations used in this study. The FLAG tag is indicated in red. (B) Localizations of pUL51 truncations in transfected Vero cells. Nuclei are stained with To-Pro3 (blue), actin stress fibers are stained with phalloidin (green) and pUL51 truncations are detected with mouse anti-FLAG (red). Representative images from three independent experiments in which >50 transfected cells were observed are shown. (C) Co-immunoprecipitation of EGFP-pUL7 with pUL51 truncations. Lysates and immunoprecipitates from Vero cells co-transfected with the indicated pUL51 constructs and with pEGFP-pUL7 plasmid are shown. The top two panels show lysate proteins detected by immunoblot using either anti-FLAG or anti- EGFP. The bottom two panels show FLAG immunoprecipitates detected with either anti-FLAG or anti-EGFP. One of two independent experiments is shown.