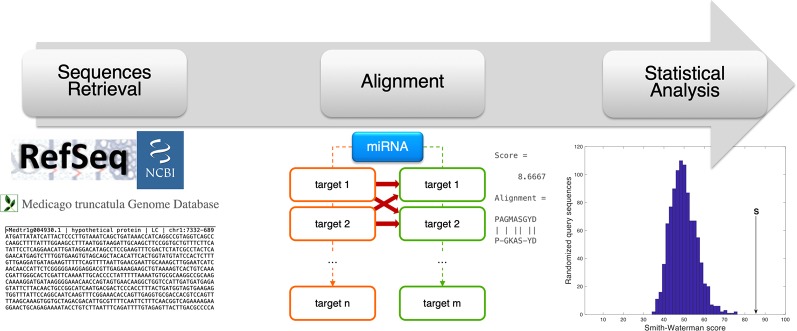
Figure 3.
Schematic representation of the alignment-based pipeline. The coding sequence (CDS) and amino acid sequence corresponding to the miRNA target genes were retrieved from online resources (RefSeq and Medicago truncatula Genome Database). For each miRNA, the CDSs and amino acid sequences of human and plant targets were compared via sequence alignment (Smith-Waterman method, by the swalign Matlab function), to compute a similarity score (provided as swalign output) for each human-plant target pair. The statistical significance of the similarity score is finally computed following a randomization method in which, for every alignment, human sequences (CDS or protein) were randomized and the distribution of swalign scores was used to compute the p-value.