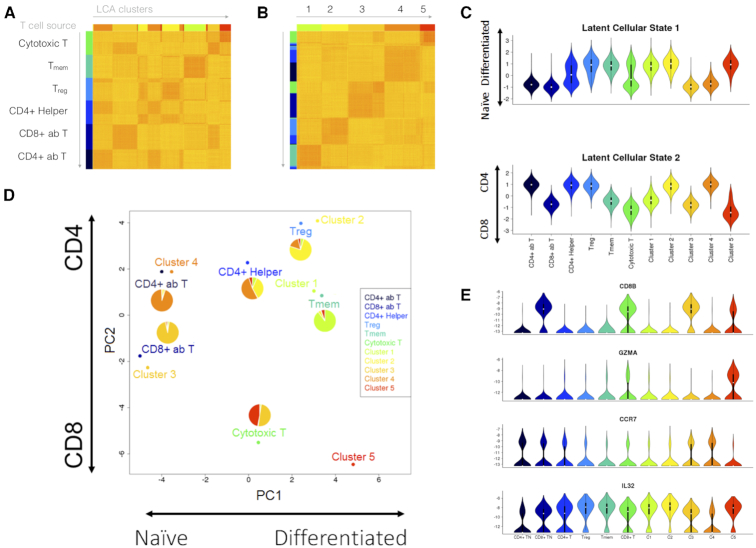
Figure 4.
Investigation of PBMC heterogeneity by using LCA. We applied LCA to data from Zheng et al. (26) for 55 000 cells representing a combination of six sorted T-cell populations. LCA identified five cell populations with biologically meaningful LC states. (A) Cell distance matrix, sorted by source ID. (B) Cell distance matrix, sorted by LCA clustering results. (C) The first LC state captured the difference between the naïve and differentiated T cells, whereas the second LC state captured the difference between the CD4+ and CD8+ cells. (D) LCA further revealed heterogeneity in the CD4+ helper T cells and CD8+ cytotoxic T cells. (E) Expression profiles of selected genes across naïve and differentiated cells in the CD4+ helper and cytotoxic T cells.