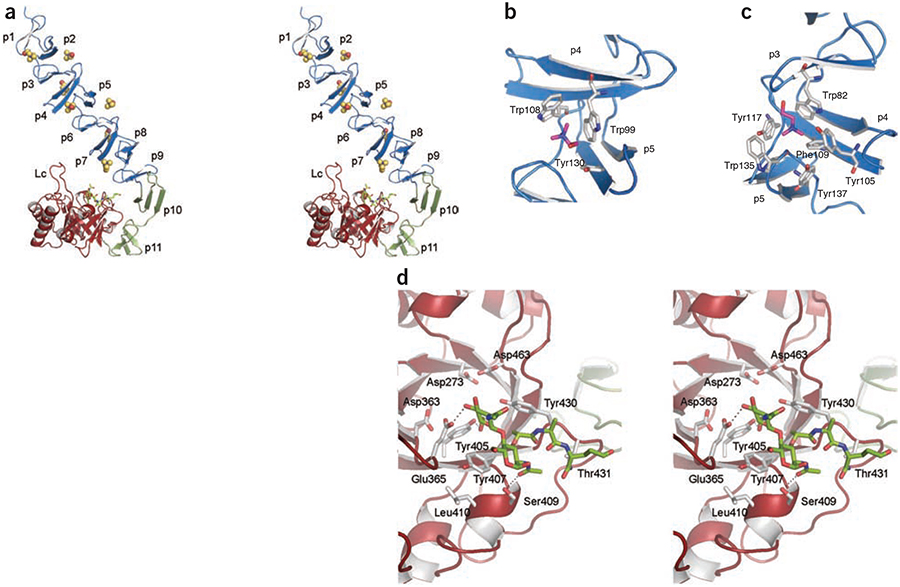
Figure 1.
3D structure of LytC–choline–PG ternary complex. (a) Stereoview representation of the general structure of LytC in complex with choline and a purified PG fragment. CBM is formed by the NI domain (repeats p1–p9) and the NII domain (repeats p10 and p11), colored in blue and green, respectively. Seven choline molecules (spheres) are bound to the choline-binding sites. The pneumococcal PG fragment (green sticks) is bound to the catalytic module (red). Loop Lc (residues 364–381) is labeled. (b) 3D structure of a choline-binding site in LytC. (c) 3D structure of GYMA sites indicates that they are formed by six aromatic residues from three consecutive repeats. In panels b and c, choline molecules and protein residues forming the cavity are depicted as capped sticks and are colored in magenta and white, respectively. (d) Details of PG recognition by LytC. The stereoview shows the interactions between LytC and a pneumococcal PG fragment. The residues forming the active site are drawn as capped sticks. Carbon atoms of the ligand are in green. Hydrogen bonds are shown as dashed lines.