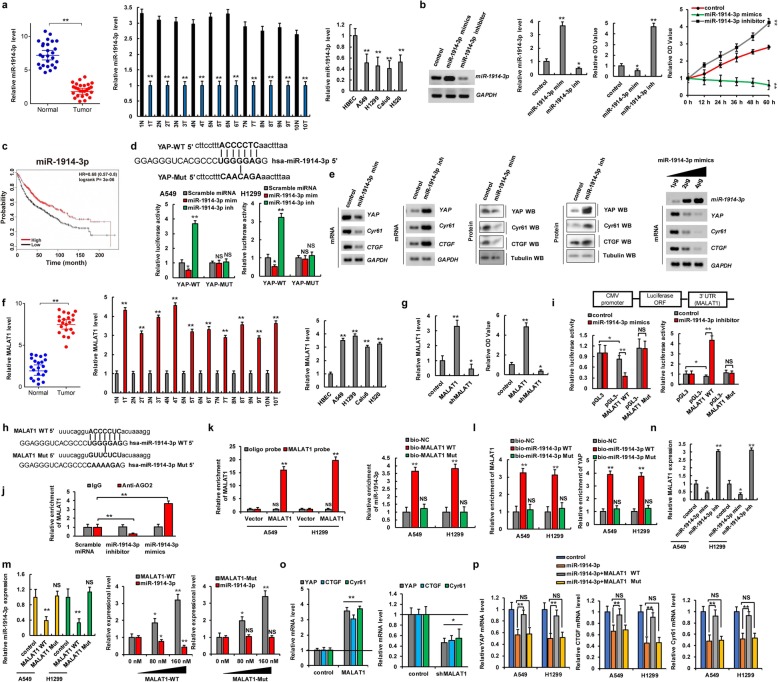
Fig. 4.
MALAT1 sponges miR-1914-3p to promote YAP expression in NSCLC. a qPCR assay indicated that the expression level of miR-1914-3p was lower in human lung cancer tissues and cell lines. b A549 cells were transfected with miR-1914-3p mimics (miR-1914-3p mim) and miR-1914-3p inhibitor (miR-1914-3p inh), respectively. The RNA level of miR-1914-3p was analyzed by RT-PCR and qPCR (left two panels). The cellular proliferation was analyzed by CCK8 assay (right two panels). c Kaplan–Meier overall survival (OS) curves of miR-1914-3p in lung cancers. d Putative miR-1914-3p binding sites in the 3′-UTR sequences of YAP (up panel). The luciferase activities were detected in A549 and H1299 cells (down panels). e The expressions of YAP, CTGF and Cyr61 were analyzed in A549 cells. f qPCR assay indicated that the RNA levels of MALAT1 were higher in human lung cancer tissues and cell lines. g The RNA level of MALAT1 was analyzed by qPCR assay (left panel). The cellular proliferation was analyzed by CCK8 assay (right panel). h Putative miR-1914-3p binding sites in the sequences of MALAT1. i The luciferase reporter activity of the wild-type and mutated MALAT1 was detected in A549 cells. j AGO2-RIP followed by qPCR to evaluate MALAT1 level after miR-1914-3p overexpression in A549 cells. k, l qPCR analyzed the RNA levels of MALAT1, miR-1914-3p and YAP in the cellular products with pull-down by biotin. m, n The RNA level of miR-1914-3p (m) and MALAT1 (n) were analyzed by qPCR. o, p The mRNA levels of YAP and its target genes, CTGF and Cyr61 were analyzed by qPCR. Results were presented as mean ± SD of three independent experiments. *P < 0.05 or **P < 0.01 indicates a significant difference between the indicated groups. NS, not significant