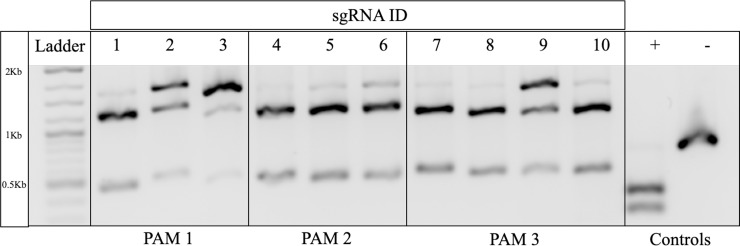
Fig 2. Results of sgRNA/Cas9 mediated cleavage of genomic DNA template.
To confirm that longer sgRNAs were capable of forming functional RNPs that are capable of creating double stranded DNA breaks in to their target gene, an in vitro cleavage assay was performed. sgRNA was combined with Cas9 enzyme and added to genomic porcine DNA that was PCR amplified around a 1600 bp region of the GGTA1 gene. The PAM site was located asymmetrically within the DNA fragment to allow for visualization of two cleaved fragments of DNA ~1200bp, ~400bp. All 10 sgRNA designed for the targeting of GGTA1 were able to form a functional RNP that targeted the GGTA1 gene. Uncleaved DNA template is shown as the largest band appearing on top of the two other fragments. The two smaller bands indicate the cleaved fragments of DNA from the sgRNA/Cas9 complex. Controls are shown on the far-right side. 2kb ladder was used for fragment size identification.