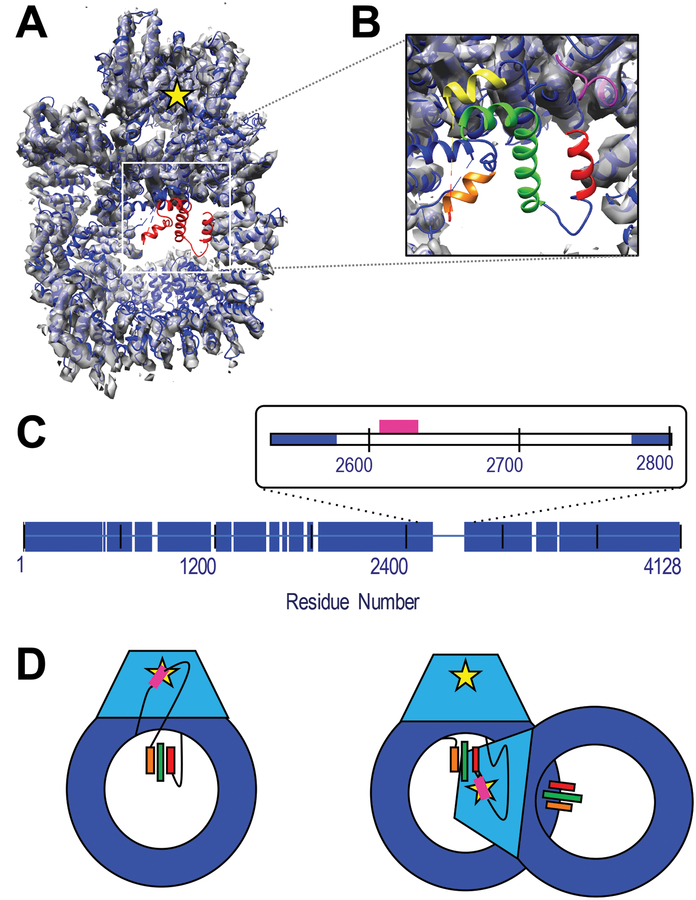
Figure 1: Structure of DNAPKcs and identification of unassigned helices.
A) The crystal structure of human DNA-PKcs (5LUQ) is shown in blue cartoon. Red cartoon identified the residues with only Cα coordinates assigned. The structure has been fitted to the 4.4 Å resolution EM map, (grey volume), (Sharif et al., 2017) which shows no density in the unassigned region. The yellow star indicates the position of the kinase active site. B) Zoom-in of unassigned region highlighting the three helices identified by DSSP as red, green and orange. The pink and yellow cartoon identifies the structured and assigned residues immediately N- and C-terminal of the disordered domain. C) Blue bars identify residues with both structure and sequence identification in the crystal structure. The inset highlights the 199-residue disordered region of interest in this study. The sector corresponding to the ABCDE cluster of phosphorylation sites (residues 2609–2638) is identified in pink. D) Cartoon representation of DNA-PKcs with the kinase domain in light blue and kinase active site as a yellow star. Potential threading arrangements that include a long linker between the ABCDE cluster (pink) and unassigned helices (colored bars) could allow an intra-molecular autophosphorylation event (left), while localization on or near the ordered helices would require a conformational change or inter-molecular autophosphorylation mechanism (right). Panels A and B were generated in part using VMD. (Humphrey et al., 1996)