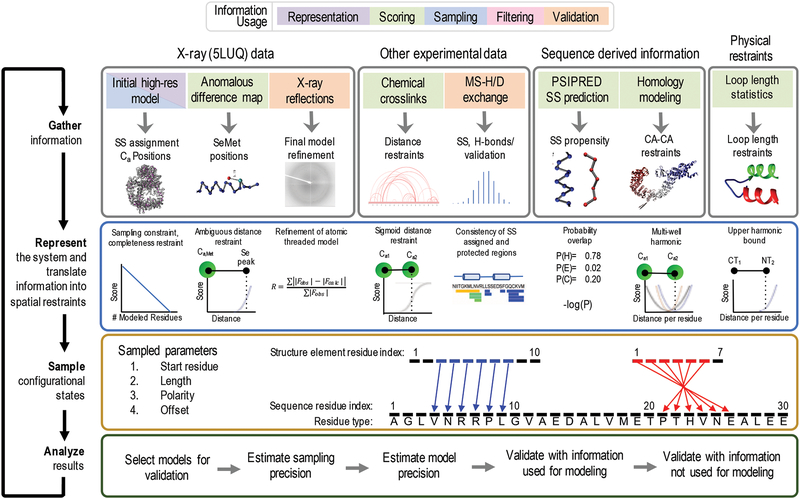
Figure 2: Four stages of integrative threading of DNA-PKcs using IMP:SSEThread.
The full integrative threading protocol proceeds through four stages. In stage 1, we gather all information about the system that we wish to use and decide at which stage of modeling we will apply it. In stage 2, we define a representation that includes the degrees of freedom we wish to assess and translate the information from stage 1 into spatial restraints [Section 2.2.2]. In stage 3, we sample alternative threading models using a Monte Carlo approach, using the scoring function from stage 2 as a guide [Section 2.2.3]. Finally, in stage 4 we assess the set of models generated in stage 3 by filtering those models that satisfy the input information, estimating the sampling and modeling precision as well as validating the models by both data used for modeling and data not used for modeling (orange boxes in stage 1).