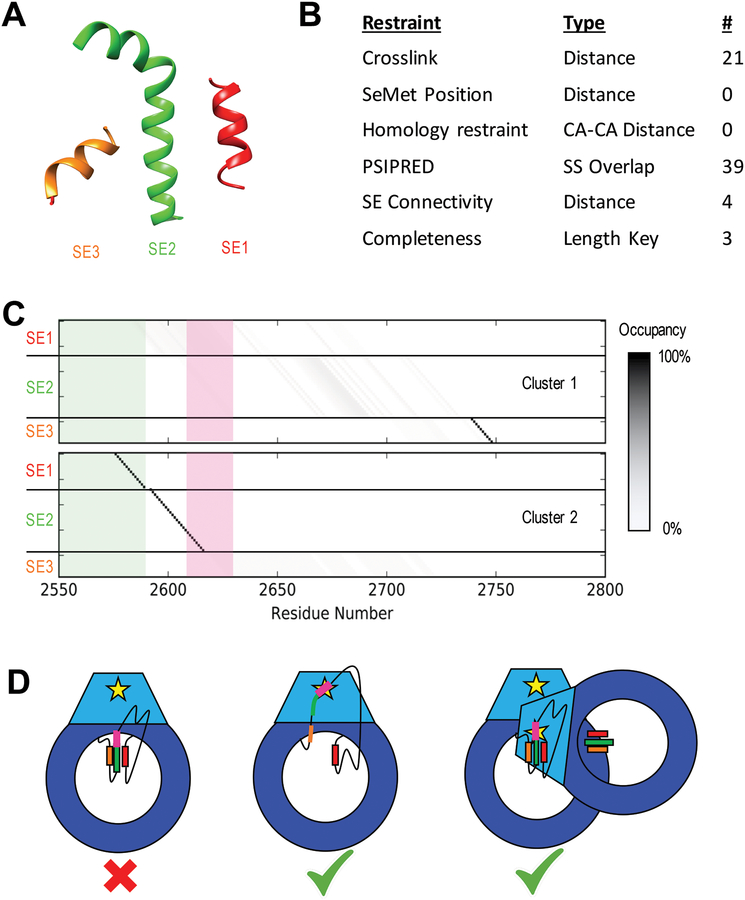
Figure 6: Results of DNA-PKcs threading.
A) Cartoon models of the three unassigned helices assigned as SEs in the DNA-PKcs crystal structure in space relative to each other (Figure 1A). B) Table of restraints utilized for SE localization using IMP:SSEThread. C) Residue occupancy of the two clusters of models identified from the top 500 threading models following enumeration of all possible start residues values. Each box represents the mapping of a residue in sequence (X-axis) to a coordinate in a structure element (Y-axis). A black box indicates that near 100% of the top models map that residue in sequence to that structure element coordinate. The green shadow highlights residues identified by hydrogen exchange as being partially protected. Cluster 1 shows a high specificity for SE3 near residue 2740 with SE1 and SE2 highly variable. Cluster 2 localizes SE1 and SE2 with high precision and has SE3 disordered. The threading solutions for SE1 and SE2 also match the HDX data, which suggest some local order in these regions. The ABCDE cluster (pink) is unlocalized in Cluster 1, while it occurs at the N-terminal end of SE2 in Cluster 2, highly constraining its position in space. D) Cartoon models of potential autophosphorylation mechanisms for the ABCDE cluster of DNA-PKcs based on Cluster 2 models. The intra-molecular mechanism (left) is not supported by this model, as the ABCDE cluster (pink) in SE2 (green) cannot interact with the kinase site (yellow star). The helices could potentially unravel (center), allowing the cluster to interact with the kinase site on the same chain. Alternatively, the other DNA-PKcs molecule in the synaptic complex could position itself to perform an inter-molecular autophosphorylation (right).