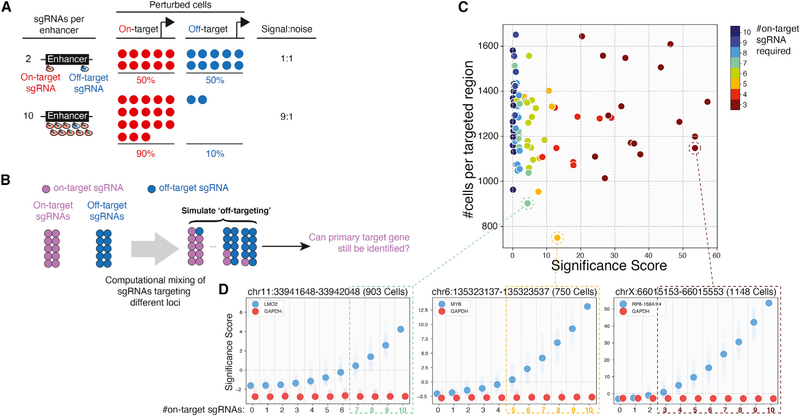
Figure 2. Increasing the Number of sgRNAs Reduces False-Positive Hits in Single-Cell Enhancer Screens.
(A) Improving signal-to-noise with more sgRNAs in single-cell screens. Increasing the number of sgRNAs targeting the enhancer increases the number of sequenced on-target cells relative to off-target cells.
(B) Overview of the computational simulation strategy. For each enhancer, we randomly discarded one to nine sgRNAs and replaced them with an equal number of random sgRNAstargeting other enhancers. Each replacement was repeated 100times, and significance scores(SS) were calculated for the top local hit of the enhancer.
(C) Scatterplot summarizing simulation results for 94 enhancers with local hits. Colors indicate the number of on-target sgRNAs required to identify the gene.
(D) Changes of SS for three example enhancers. Solid dots represent the median of 100 iterations. Transparent dots represent all iterations. Dashed boxes indicate parameters allowing known target genes to be identified (SS > 0).
See also Figure S2.