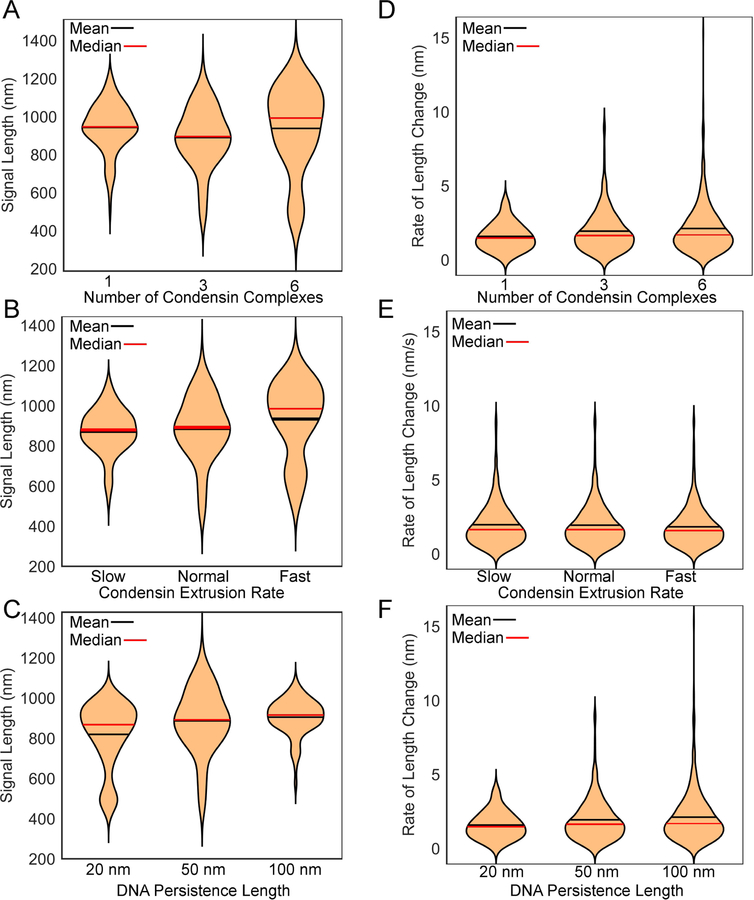
Figure 7. Violin plots of dicentric plasmid simulations.
Violin plots comparing the simulated tetO/TetR-GFP signal lengths in plasmid simulations with differing numbers of condensin complexes (A), extrusion rates (B), and DNA persistence lengths (C). Violin plots comparting the rate of change in the lengths of the simulated tetO/TetR-GFP signal lengths in plasmid simulations with differing numbers of condensin complexes (D), extrusion rates (E), and DNA persistence lengths (F). Unless otherwise indicated plasmids have 3 condensin complexes, a normal extrusion rate, and the DNA has a persistence length of 50 nm.