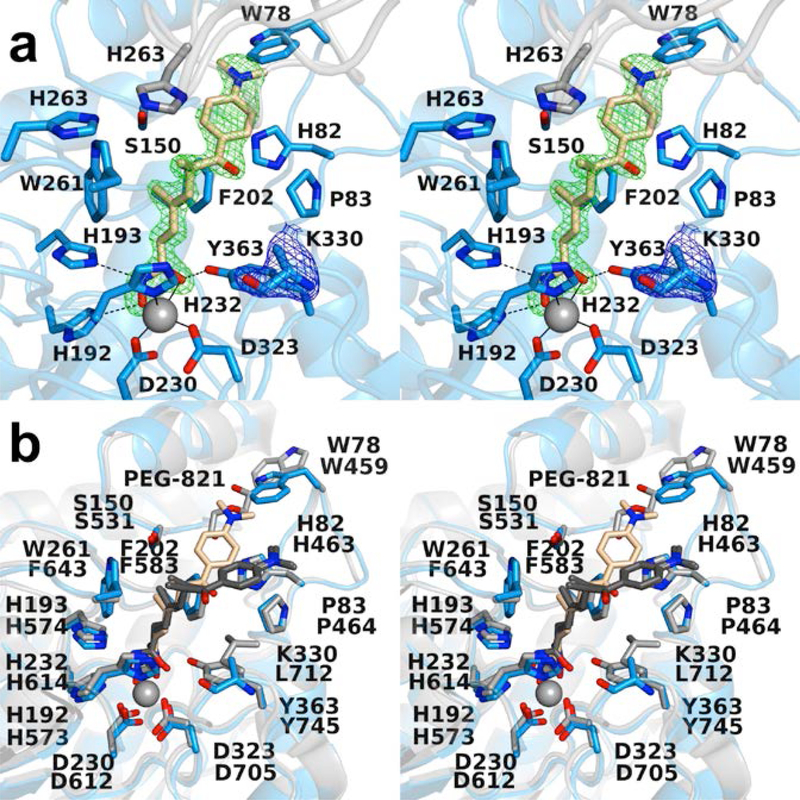
Figure 2.
(a) Polder omit maps of the HDAC6 CD1–Trichostatin A complex (PDB 6UO2; monomer B, inhibitor contoured at 5.5 σ; K330 is contoured at 3.5 σ). Atoms are color-coded as follows: C = light blue (monomer B), light gray (monomer A), or wheat (inhibitor), N = blue, O = red, and Zn2+ = gray sphere. Metal coordination and hydrogen bond interactions are indicated by solid and dashed black lines, respectively. (b) Superposition of the HDAC6 CD1–Trichostatin A complex (monomer B, color-coded as in (a)) and the HDAC6 CD2–Trichostatin A complex (PDB 5WGI; monomer A, C = light gray (protein) or dark gray (inhibitor). W78 of CD1 is conserved as W459 in CD2, and this residue adopts alternate conformations in CD1 (“in”) and CD2 (“out”). The “out” conformation of W459 at the mouth of the HDAC6 CD2 active site enables the binding of polyethylene glycol fragment PEG-821.