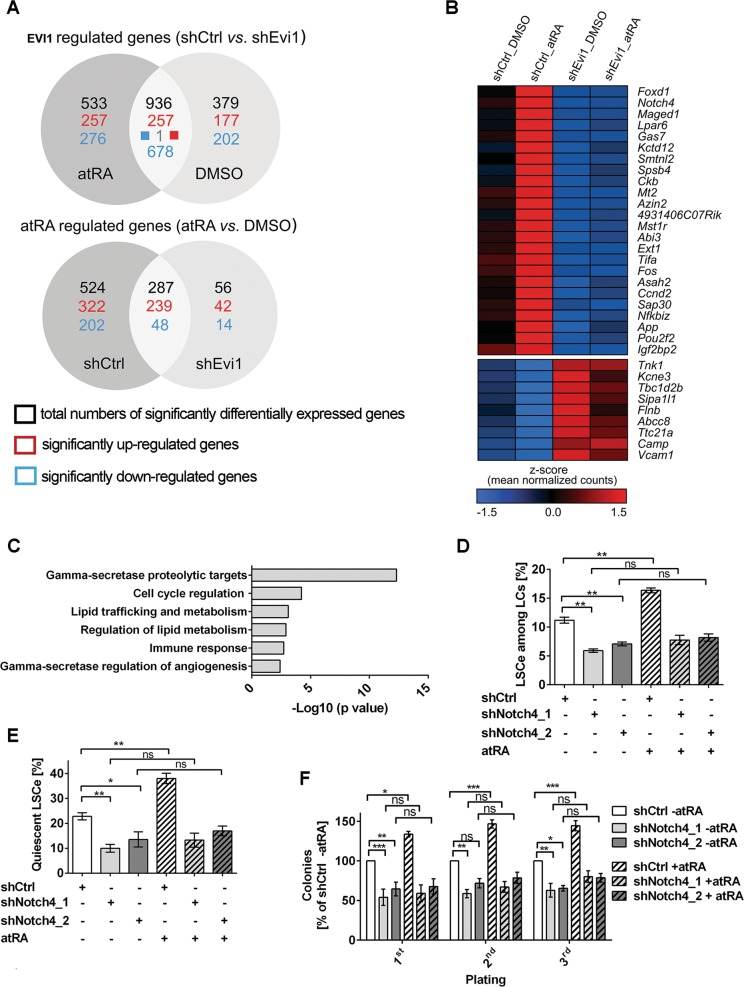
Fig. 4. EVI1 augments transcriptional responses to atRA in LSCe, and Notch4 is a relevant joint target.
LSCeLSK_MA9_shCtrl and LSCeLSK_MA9_shEvi1 were treated with atRA or solvent for 24 h and subjected to RNA-sequencing. a Numbers of genes regulated by EVI1 or atRA at an FDR < 0.05. b Heatmap of genes that were minimally affected by atRA in LSCeLSK_MA9_shEvi1, but whose regulation by EVI1 was enhanced by atRA in LSCeLSK_MA9_shCtrl. c Pathway enrichment analysis of genes shown in b. d–f BM cells from mice terminally ill after transplantation with MA9 transduced LSK cells were further transduced with shCtrl, shNotch4_1, or shNotch4_2. Venus+ RFP+ cells were treated with 1 µM atRA or an equivalent amount of solvent for 3 days. n = 3; *p < 0.05; **p < 0.01; ***p < 0.001; ns, not significant; ANOVA followed by Bonferroni's post-hoc test. d Proportions of LSCe among LCs. e Proportions of quiescent LSCe (LSCe in G0). f Colony formation in methyl cellulose, presented as percent of solvent-treated, shCtrl transduced LCLSK_MA9 in each round of plating.