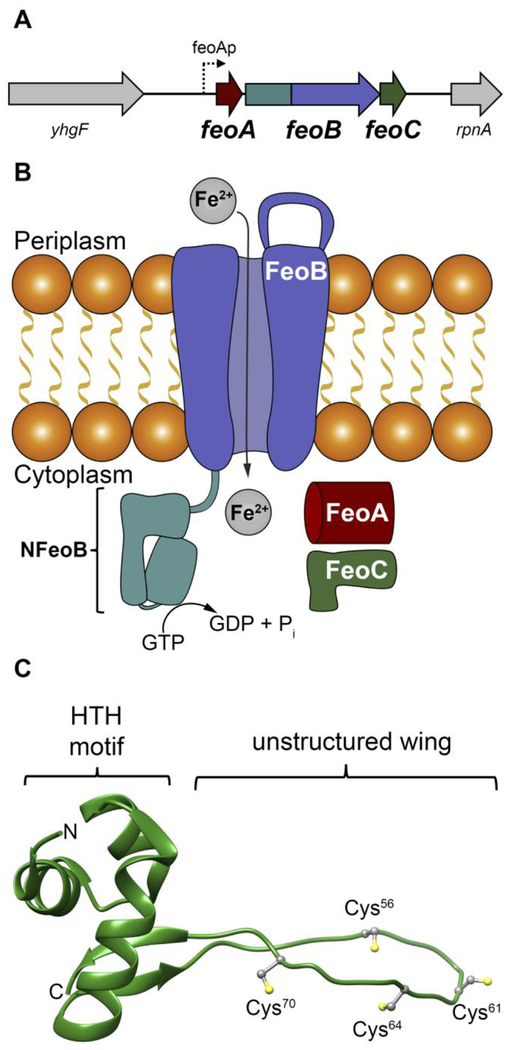
Figure 1.
The Feo system and the structure of E. coli FeoC. A. The arrangement of the feo operon in E. coli K-12, which encodes for three proteins: FeoA, FeoB, FeoC. FeoAp represents the location of the FeoA promoter. To emphasize the co-transcription of the components of the feo operon, the physical layout of neighboring genes such as a putative RNA-binding protein (encoded by yhgF) and a downstream nuclease (encoded by rpnA) is included. B. Cartoon of the Feo system in E. coli. FeoA (red) and FeoC (green) are small cytosolic proteins that may function as regulatory accessories to control ferrous (Fe2+) iron transport. Movement of ferrous iron across a cellular membrane is accomplished by the large, polytopic membrane protein FeoB (purple). Hydrolysis of GTP to GDP within the N-terminal soluble GTP-binding domain of FeoB (NFeoB, teal) is thought to regulate opening and closing of FeoB, but it is unknown whether this process is driven in an active or facilitated manner. C. Lowest-energy NMR conformer of EcFeoC (PDB ID 1XN7). Labeled regions are: the helix-turn-helix (HTH) motif and the unstructured wing region that contains four Cys residues (Cys56, Cys61, Cys64 and Cys70) involved in [Fe-S] cluster binding. The labels “N” and “C” represent the amino and carboxy termini, respectively.