Figure 2 –
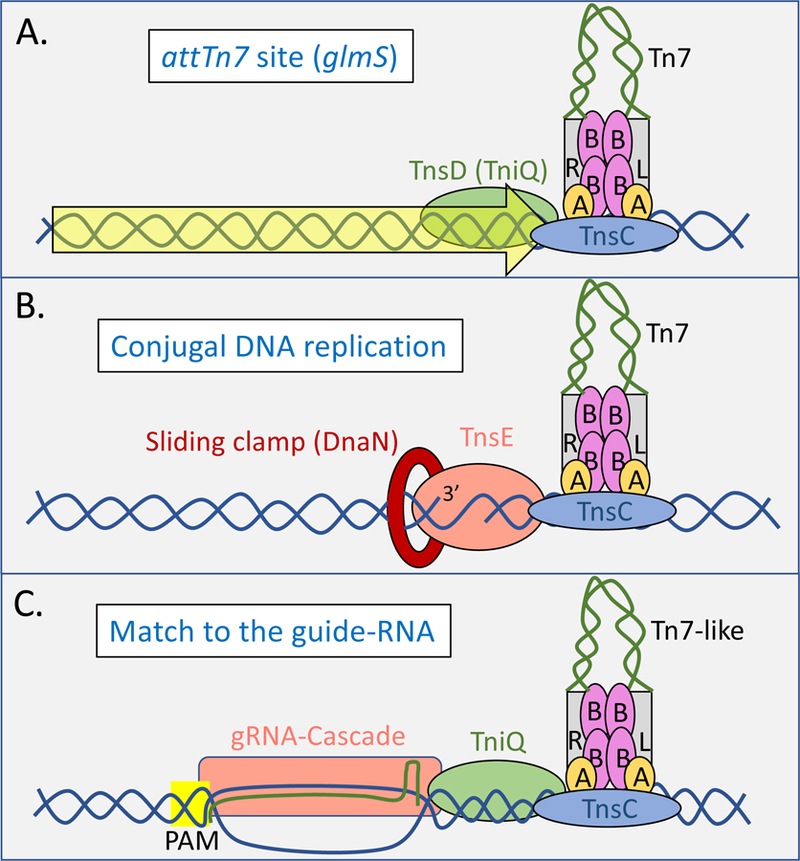
Models for how target complexes are assembled at various preferred target sites with Tn7 and Tn7-like elements. The Left (L) and Right (R) ends of the Tn7/Tn7-like elements are indicated with grey rectangles and the body of the element in green. The ends of the element are synapsed with TnsB (purple ovals, labeled B) and shown interfaced at the blue target DNA with TnsC (blue oval) and TnsA (yellow ovals, labeled A) to the site of insertion as recruited by three different targeting mechanisms. A. Prototypic Tn7 transposition into the attTn7 site uses the sequence specific DNA binding protein TnsD(TniQ) (green oval) that recognizes the 3’ coding region of the glmS gene (yellow block arrow) to recruit and activate transposition. Insertion occurs at a single position with the right end of the element integrated closest to glmS. Tn7-like elements may use a similar process for recognizing other att sites using TniQ as described in the text. B. Prototypic Tn7 transposition that recognizes active DNA replication found during plasmid conjugation uses the TnsE proteins which recognizes two features of discontinuous DNA replication in recipient cell bacteria; interaction with the sliding clamp (DnaN – red ring) and 3’ recessed-end substrates to direct transposon integration. Tn7 insertion events occur at multiple positions, but in a single origination with the direction of discontinuous DNA replication in recipient bacteria. C. Tn7-like elements with the I-F variant CRISPR/Cas system use a guide RNA(gRNA) complex to recognize a protospacer with the correct PAM sequence as predominantly found in mobile plasmids to direct transposition events ~49 bp from the protospacer with the right end of the element proximal to the end of the protospacer through an association with TniQ.
