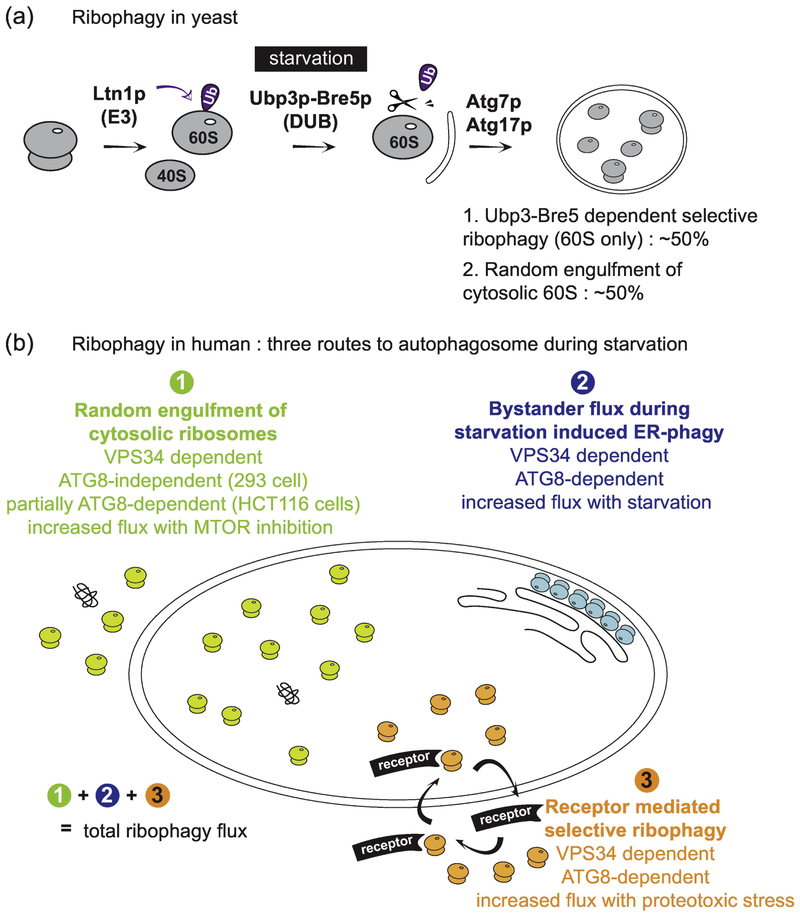
Fig. 3.
Overview of proposed ribophagy pathways in yeast and mammals. (A) During nutrient rich conditions, it has been reported that ribophagy in yeast is protected by the ubiquitylation of Rpl25 by the E3 ubiquitin ligase Ltn1. Upon starvation, the deubiquitylation enzyme Ubp3p-Bre5p removes the ubiquitin from Rpl25p, which subsequently leads to the selective sequestration of the 60S particle by autophagic membranes through an unknown mechanism. Ubp3p-Bre5p-dependent ribophagic flux is responsible for ~50% of the total ribophagic flux during starvation, but there is still significant flux (~50%) in Ubp3Δ cells, suggesting that there may be a pool of ribosomes that is differentially regulated. Whether or not an unknown receptor protein mediates the binding of 60S ribosomes to ATG8, and how the 40S assembly is selectively targeted is not known. (B) Ribosomes in mammals can be encapsulated in autophagosomes by three independent routes: 1) random sequestration during bulk autophagy, 2) by-stander flux during ER-phagy, and 3) receptor-mediated selective enrichment into autophagosomes. Quantitative methods and flux measurements are required to understand the contribution of each pathway under specific conditions.