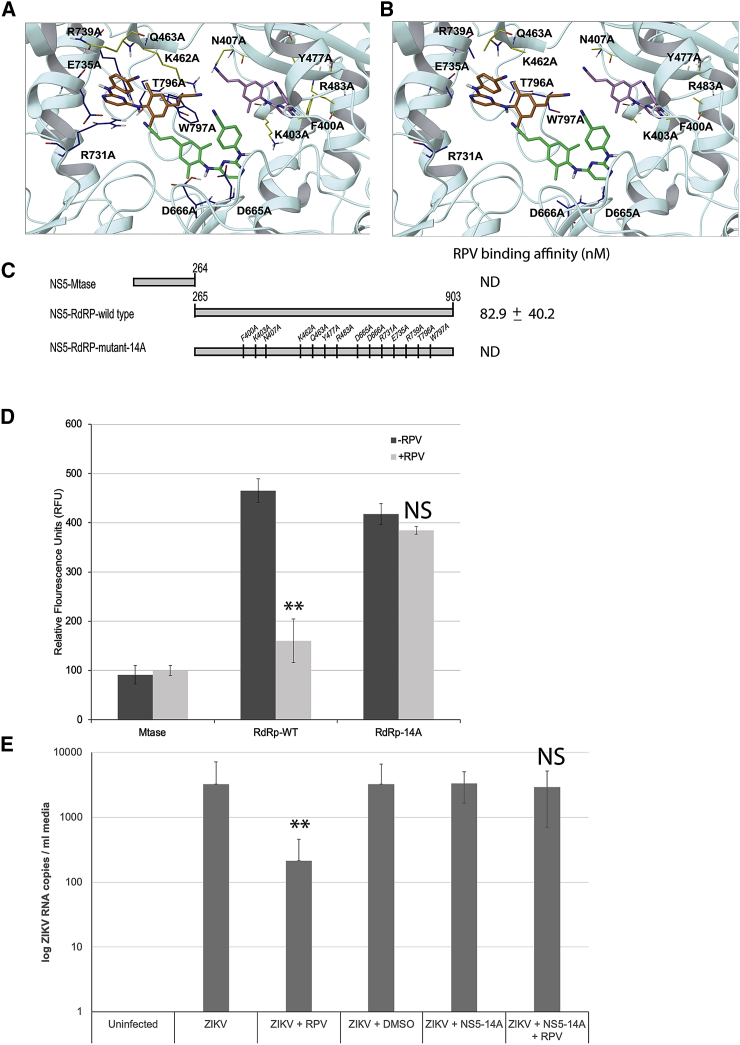
Figure 4.
Physical and Functional Assessment of RPV Binding to NS5 Mutants with Site-Directed Mutagenesis
Structural features of ZIKV NS5-RdRp wild-type (A) and NS5-RdRp mutant-14A (B) and their potential interaction with RPV were characterized via computation. In both wild-type (A) and mutant-14A (B) panels, side chains are color coded by NS5 mutant (yellow or blue for mutant-N7A or mutant-C7A, respectively). Binding modes are ranked by docking score (kcal/mol): −7.4 (best predicted binding), −6.6, and −5.6 for RPV poses in green, orange, and pink carbons, respectively. The bottom ranking pose (pink carbons) is obtained for RPV interacting with mutant-N7 residues and shows no binding; hence, this binding mode (pink carbon RPV) is less likely to occur. (C) Schematic presentation of amino acid mapping of NS5 wild-type and side-directed mutations (14A) are illustrated. The relative fluorescein emitted by the thermal shift dye is recorded during the temperature ramp phase and plotted versus temperature. ND denotes no significant affinity detection. (D) Effect of RPV on RNA polymerase activities of MTase domain and RdRp wild-type and RdRp-14A proteins was also analyzed by RdRp assay. 1 μg ZIKV RNA and 500 ng recombinant NS5 full-length or mutant proteins were added to the reaction buffer in the presence and absence of RPV (10 ng) and analyzed as described in the Materials and Methods. NS5 activity was calculated from three independent experiments and quantified as relative fluorescence units (RFU) emitted during the reaction. (E) ZIKV RdRp-14A mutant was cloned into a mammalian expression vector. PHFA cells were transiently transfected with this construct and either treated with RPV or left untreated. Cells were infected with ZIKV (0.1 PFU) at 24 h post-transfections, and growth media of cells were analyzed by qRT-PCR for ZIKV RNA copies. **p < 0.05.