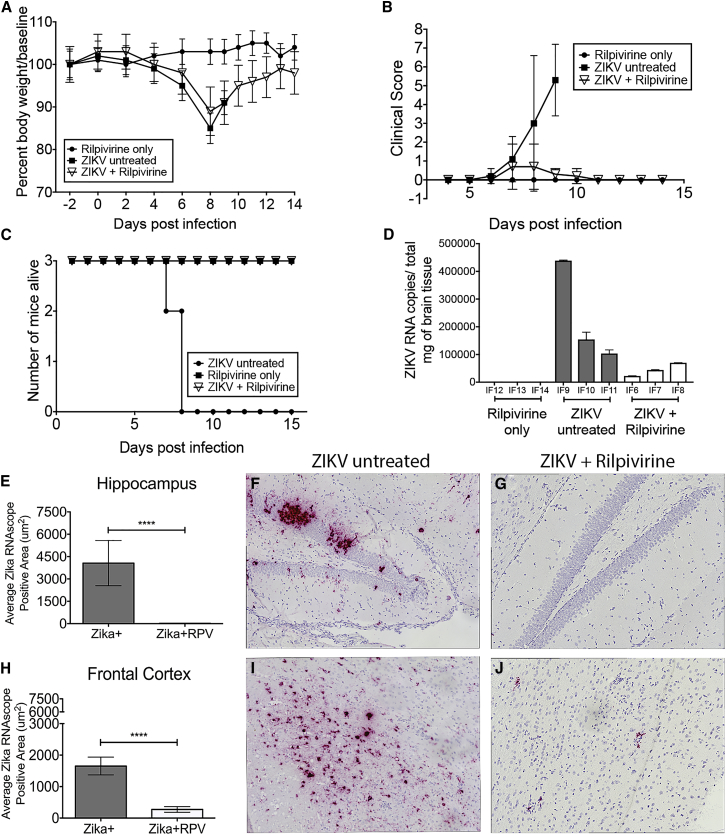
Figure 5.
RPV Inhibits ZIKV Propagation, Reverses the Mortality, and Decreases the Viral Burden in the Brain of IFNR−/− Mice
(A) Body weights were measured for each mouse from the three groups (closed circle, rilpivirine only; closed square, ZIKV untreated; and open triangle, ZIKV + RPV) until the end of the study (14 days post-infection [dpi]). Data are shown as a percent of body weight compared to baseline. (B) Grasp tests were performed daily starting at 4 dpi and analyzed for clinical scoring for the three groups. (C) Survivor curve analysis is shown based on the Kaplan-Meier estimates. All animals in the uninfected + RPV and ZIKV-infected + RPV groups survived until the end of the study. The ZIKV-infected and untreated mice were sacrificed at 7 and 8 dpi. (D) ZIKV RNA copies per milligram total brain lysates are presented per animal (IF6–IF14). The mean and SD are shown, and the SD was calculated from triplicate analysis of the RNA samples from each mouse. (E–J) ZIKV RNA was visualized with RNAscope (red) and quantified in the hippocampus and the frontal cortex of ZIKV-infected untreated animals (ZIKV+) and ZIKV-, RPV-treated animals (ZIKV + RPV). (E) The average ZIKV+ RNA area in the hippocampus of the ZIKV-untreated (gray bars) mice compared to the ZIKV + RPV group (white bars) showed a significant reduction in the ZIKV RNA with RPV (p < 0.0001). (F) Representative image of hippocampus ZIKV RNAScope staining reveals high viral RNA signal in ZIKV mice. (G) Representative image of hippocampus ZIKV RNAScope staining shows significantly less signal in ZIKV + RPV mice. (H) The average ZIKV+ RNA area in the frontal cortex of the ZIKV-untreated (gray bars) mice compared to the ZIKV + RPV group (white bars) showed a significant reduction in the ZIKV RNA with RPV (p < 0.0001). (I) Representative image of a frontal cortex ZIKV RNAScope in situ reveals high viral RNA signal in ZIKV mice. (J) Representative image of frontal cortex ZIKV RNAScope staining shows significantly less signal in ZIKV + RPV mice. The average ZIKV RNA signal area was quantitated as the average of 10, non-overlapping 40× images from each animal (n = 3/group), using a Keyence BZ-X700 microscope. Bar graphs show average ZIKV RNA area (mean, SEM). Comparison of means was determined by Mann-Whitney, two-tailed t test (*p < 0.05, ****p < 0.0001).