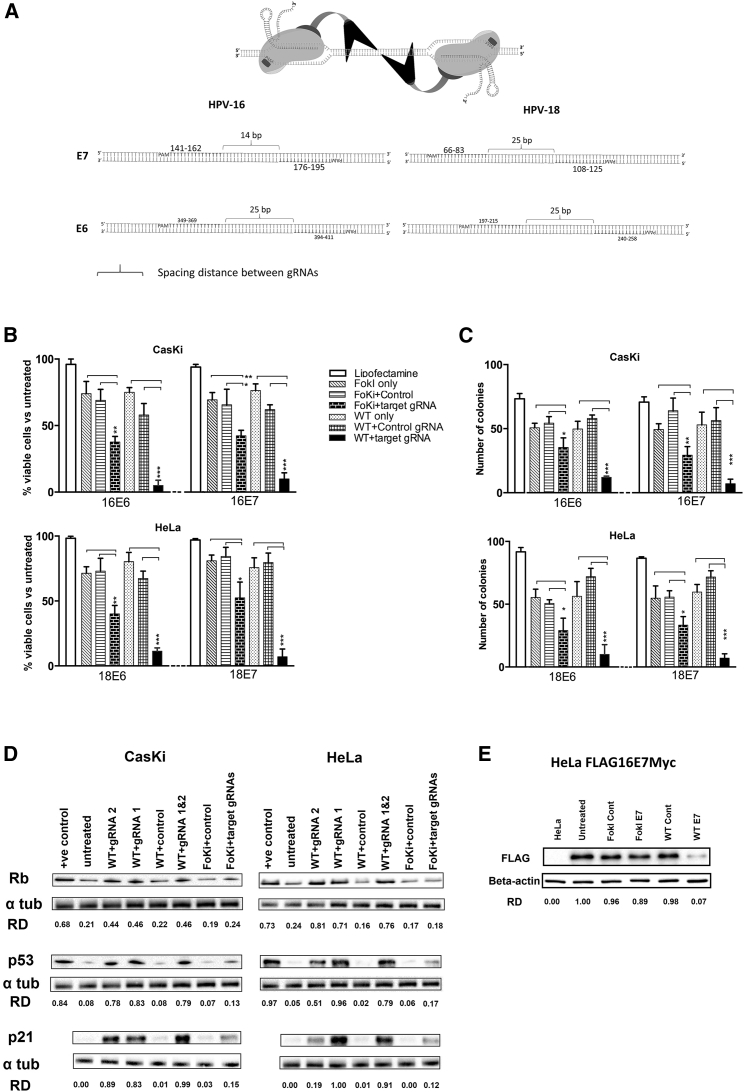
Figure 1.
CRISPR/Cas Editing of the HPV E6 and E7 Genes Reduces Cell Viability and Induces Key Growth Control Proteins
(A) The design strategy and the binding sites for gRNAs against HPV E6 and E7 genes for both dCas9-FokI and WT Cas9 variants. (B) Cells (CasKi, HPV16 positive; HeLa, HPV18 positive) were treated with target-specific gRNAs (16E6, 16E7, 18E6, or 18E7) or control gRNA (non-specific), co-transfected with either the WT Cas9 or FokI-dCas9 for 72 h before cell viability was determined using MTT assay. Viability was expressed as a percentage relative to the untreated group (data not shown). (C) Cells (CasKi, HPV16 positive; HeLa, HPV18 positive) were treated with target-specific or control gRNAs with either WT Cas9 or FokI-dCas9, and then allowed to form colonies over 2 weeks before the number of colonies was counted. (D) CasKi (HPV16 positive) or HeLa (HPV18 positive) cells were treated with target-specific or control gRNAs and either WT Cas9 (WT) or FokI-dCas9 (Foki) for 72 h before protein expression of various growth control proteins (p53 and p21 for E6 targeting, Rb for E7 targeting) was determined by western blot analysis. The effect of WT Cas9 with target-specific gRNA 1 (WT 1) or gRNA 2 (WT 2) or both (WT 1 and 2, 1:1 ratio) was tested (Table S1 for the list of gRNAs). The relative density (RD) of bands was quantified using ImageJ software. Jurkat and HEK293 lysates were used as positive control for Rb and p53, respectively. (E) HeLa Flag16E7Myc cells were treated with 16E7 gRNAs and WT Cas9 or FokI-dCas9 before protein expression of FLAG-tagged E7 was quantified by western blotting. HeLa cell lysate was used as a negative control. All data are presented as mean ± SD. Statistical difference was assessed by ANOVA with post hoc analysis, *p < 0.05, **p < 0.01, ***p < 0.001.