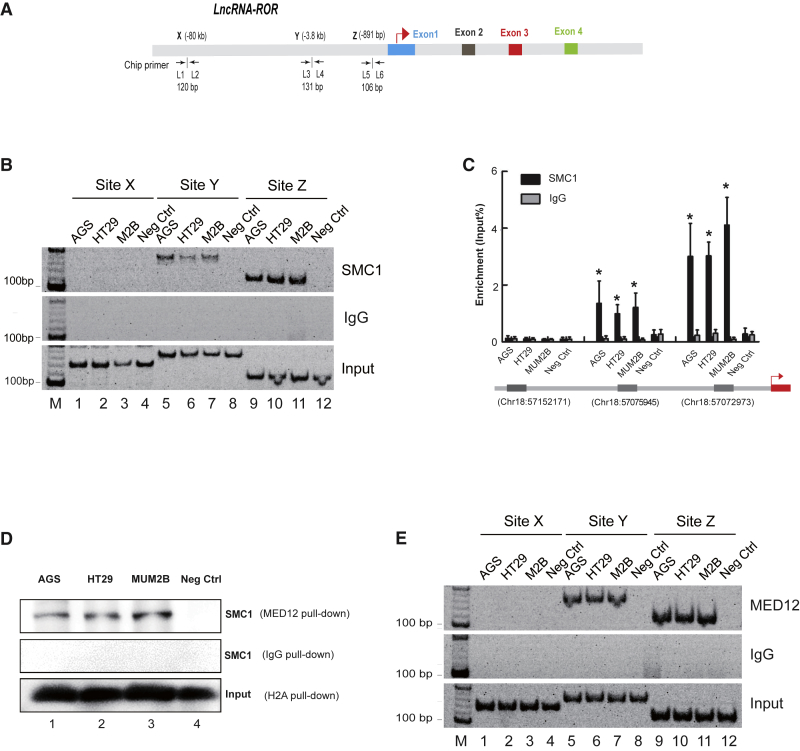
Figure 2.
SMC1 Forms Intrachromosomal Looping at the ROR Locus
(A) Schematic diagram of the ROR lncRNA and the position of primers used for ChIP analyses. L1 through L6, primer names; arrows, transcriptional direction. (B) The interaction between SMC1 and ROR lncRNA. ChIP assay demonstrating that SMC1 binds the ROR promoter (−891 bp) and the upstream fragment (−3.8 kb). IgG is used as a negative control. Sites X, Y, and Z are ChIP detection sites. (C) The qPCR-ChIP assay showing the interaction between SMC1 and the ROR lncRNA. ChIP sites on the chromosome are listed at the bottom. Immunoglobulin G (IgG) is the negative control. All of the data are presented as the mean ± SD. *p < 0.05 compared with the negative control (fibroblast). (D) SMC1 interacted directly with MED12 as detected by co-immunoprecipitation. Cohesin (SMC1) is detected by western blot after AGS, HT29, and MUM2B immunoprecipitation. (E) ChIP assay detecting the interaction between MED12 and the ROR lncRNA. Sites X, Y, and Z are ChIP detection sites. M, marker.