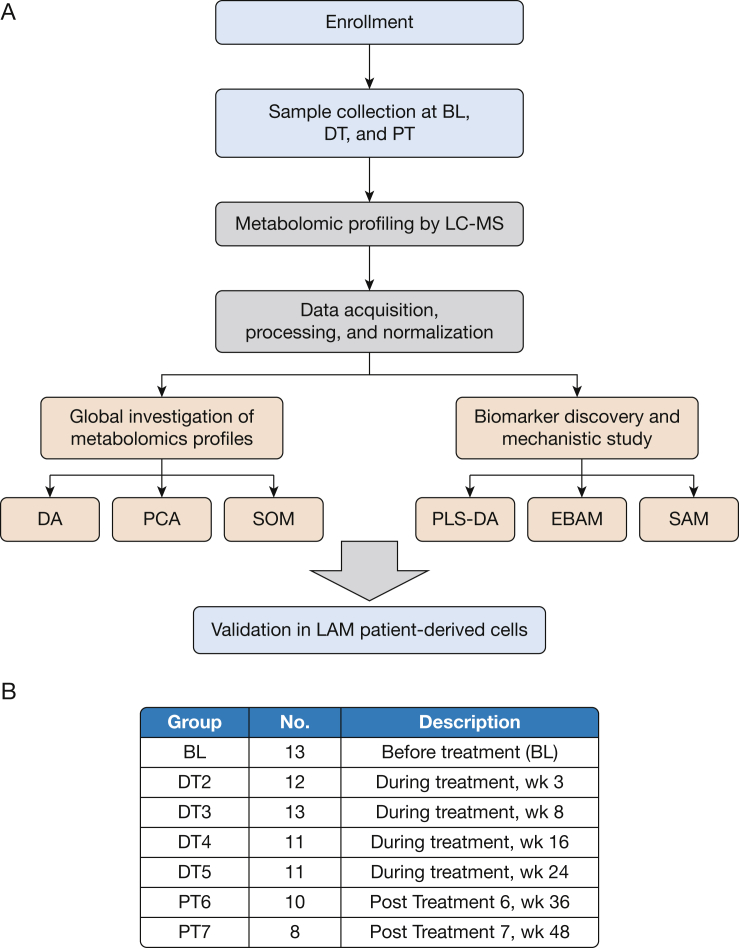
Figure 1.
Overview of data collection and analysis. A, Sample collection and analysis pipeline. Plasma samples were collected before treatment as baseline reference, during a 24-week treatment period consisting of four visits, and from the posttreatment observation period consisting of two visits. Metabolomic profiling was performed by liquid chromatography-mass spectrometry. Principal component analysis and a self-organizing map were used for global metabolomic profiles analysis. Differential analysis was used for identification of changes of metabolite levels caused by treatment. Three statistical models were employed for biomarker identification: partial least squares-discriminant analysis, empirical Bayes analysis of microarrays, and significance analysis of microarrays. Identified biomarkers and metabolic pathways were validated in a TSC2-deficient cell line (621-101) derived from a patient with lymphangioleiomyomatosis. B, Summary of sample collections. Thirteen subjects were enrolled in the study and provided baseline plasma samples. During the period of treatment, plasma samples were collected at up to four visits, resulting in a total of 47 samples. In total, 18 samples were collected at two posttreatment visits. BL = baseline; DA = differential analysis; DT = during treatment; EBAM = empirical Bayes analysis of microarrays; LAM = lymphangioleiomyomatosis; LC-MS = liquid chromatography-mass spectrometry; PCA = principal component analysis; PLS-DA = partial least squares-discriminant analysis; PT = posttreatment; SAM = significance analysis of microarrays; SOM = self-organizing map.