Figure S3.
In Vitro Recreation of Hopx+ CARSCs and Its Regenerative Potential, Related to Figure 3
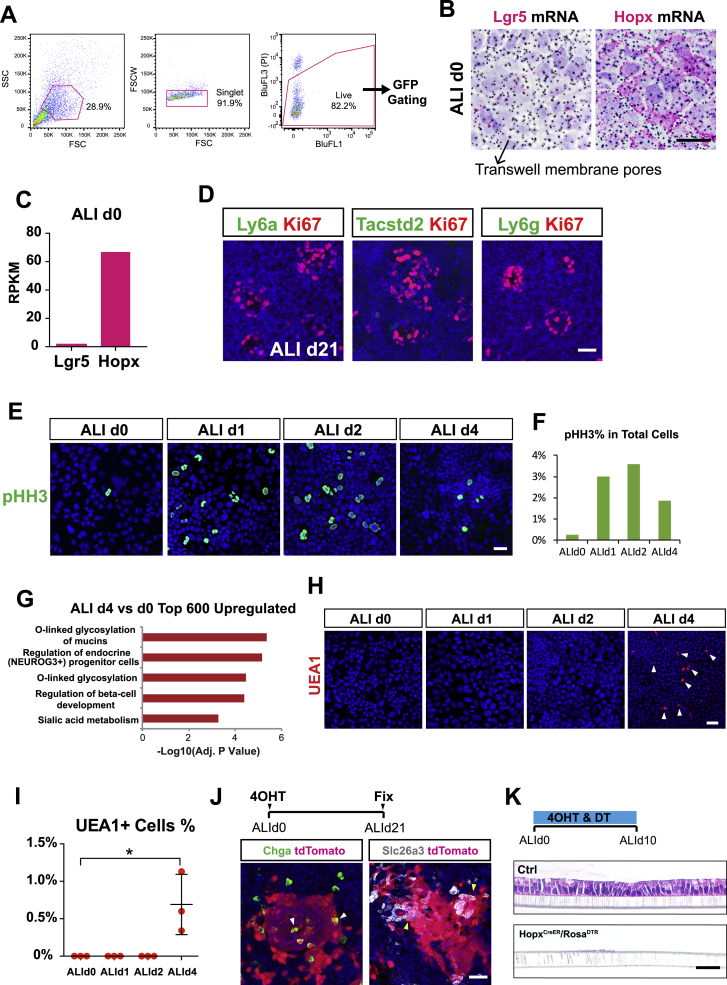
(A) Detailed flow cytometry gating strategies used to select live single cells for subsequent GFP analysis in WT, Lgr5GFP and HopxGFP ALId0 cells as shown in Figure 3B. SSC = Side scatter. FSC = Forward scatter. BluFL1 was used to check for autofluorescence of the BluFL3 (propidium iodide, PI) channel which is a live/dead indicator.
(B) Whole mount in situ hybridization on Transwells to assay Lgr5 and Hopx mRNA levels in ALId0 monolayers (red). Black dots represent the pores of the Transwell membrane.
(C) Expression of Lgr5 and Hopx mRNAs on ALId0 as defined by RNA-Seq.
(D) Whole mount Ki67 (red) co-staining with Ly6a (green, left), Tacstd2 (green, middle) and Ly6g (green, right) on ALId21 monolayers.
(E-F) Whole mount phospho-histone H3 staining on Transwells examined on ALId0, ALId1, ALId2 and ALId4. The mitotic rate for these time points was plotted in panel F.
(G) Top KEGG pathways upregulated in ALId4 monolayers in comparison to ALId0.
(H-I) Whole mount UEA1 staining of Transwells sampled on ALId0, ALId1, ALId2 and ALId4 to characterize the time course of goblet cell differentiation. The percent of UEA1+ cells is indicated in (I). n = 3 samples/group.
(J) Whole mount co-staining of Chga and Slc26a3 with tdTomato following 21 days of lineage tracing experiment on ALI cultures derived from HopxCreER/RosaTd mice.
(K) H&E staining of ALI culture sections after 10 days of Hopx+ cell ablation.
Mean values ± SD are shown. Two-tailed Student’s t test: ∗p < 0.05. Quantification data are represented as mean ± SD. Bars: (B, D, E, H, J and K) 25 μm. In situ hybridization and immuno-fluorescent images are representative of at least three samples examined.